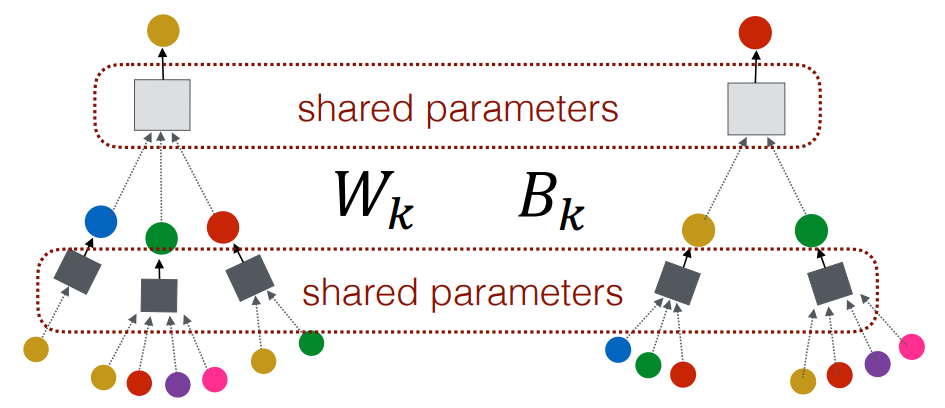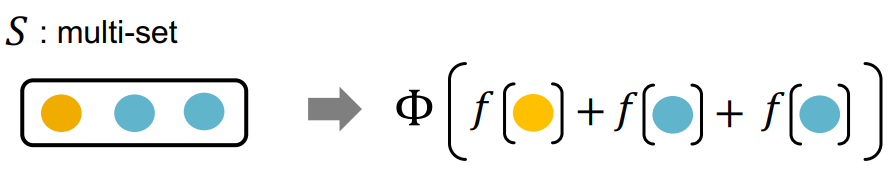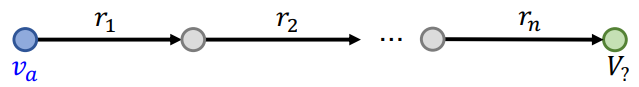Graph
Definition
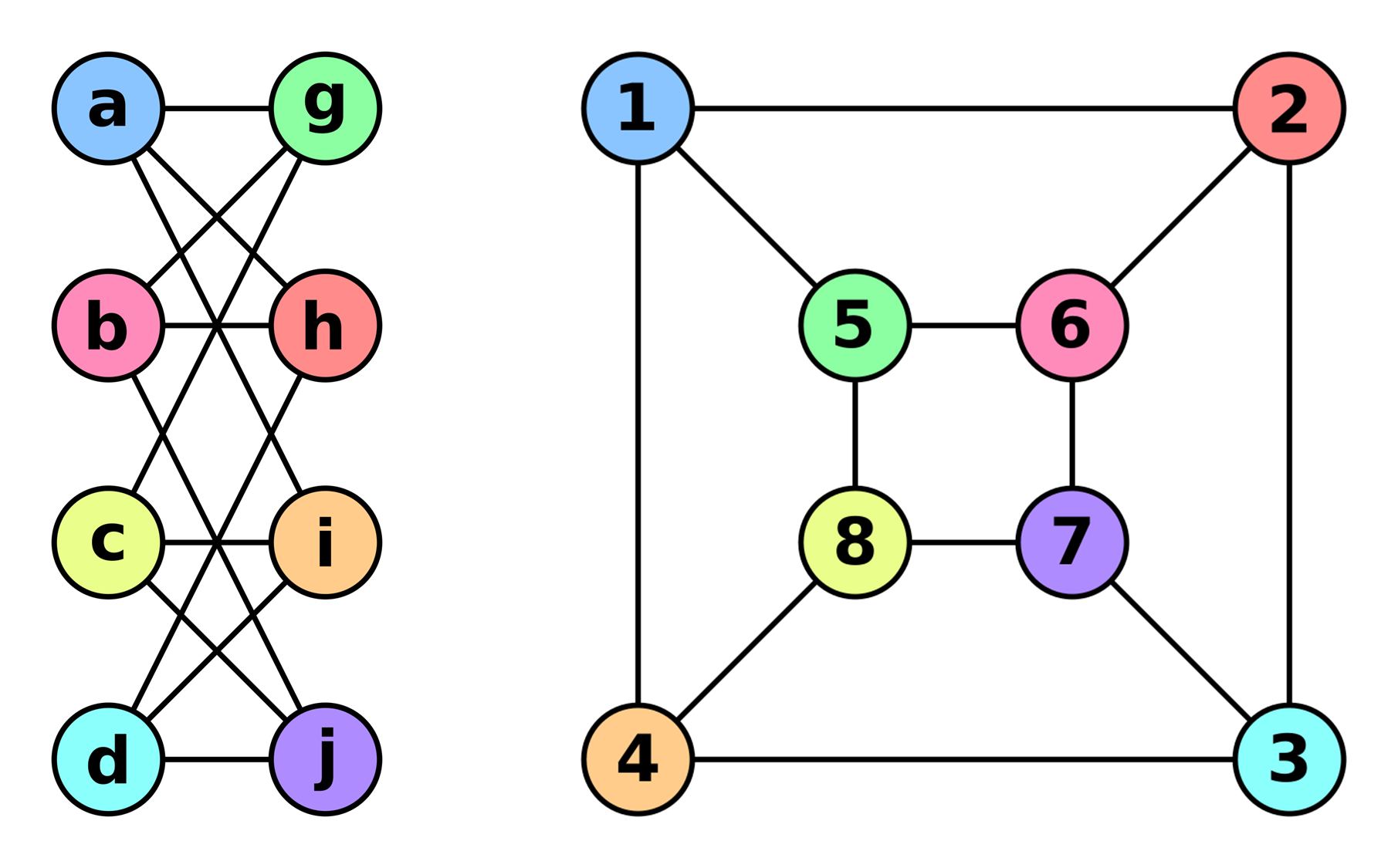
A graph is a pair of a set of vertices (nodes) and a set of pairs of vertices (edges) .
Link to original
Node Embeddings
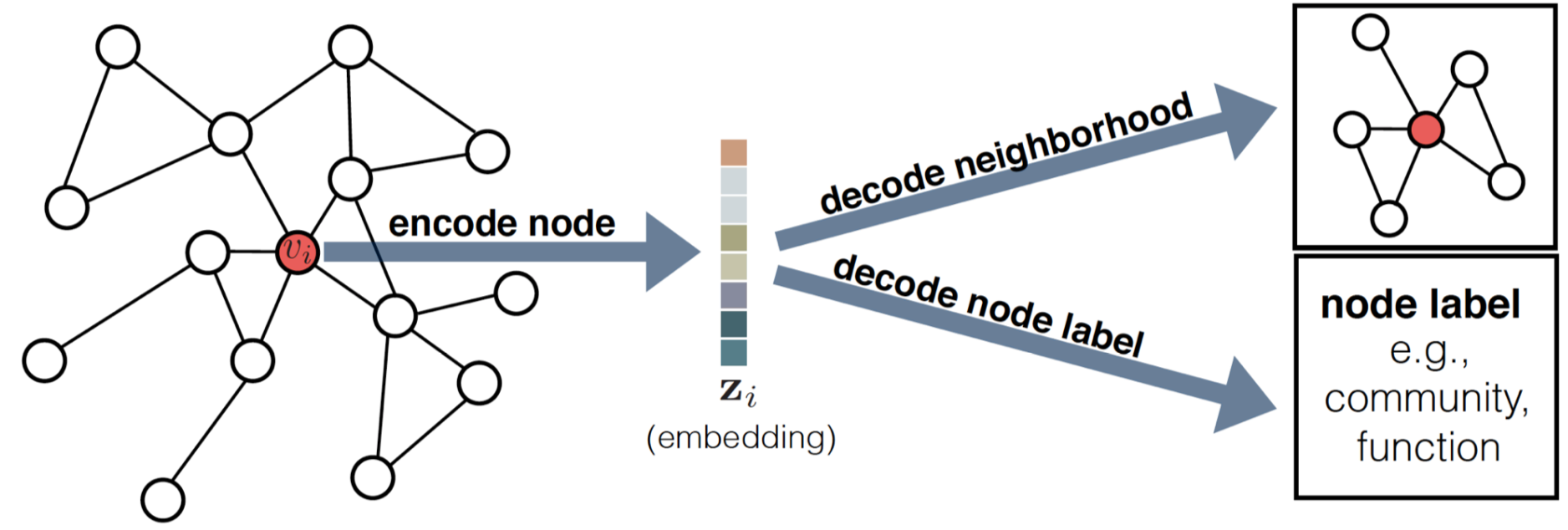
Node Embedding
Definition
The goal of node embedding is finding an encoder mapping nodes into embedding space preserving the similarity of the nodes.
The encoder maps node to embedding and the decoder extracts useful-information such as local neighbor and classification label of the corresponding node from embedding, where is the dimension of the embedded vectors. .
Link to original
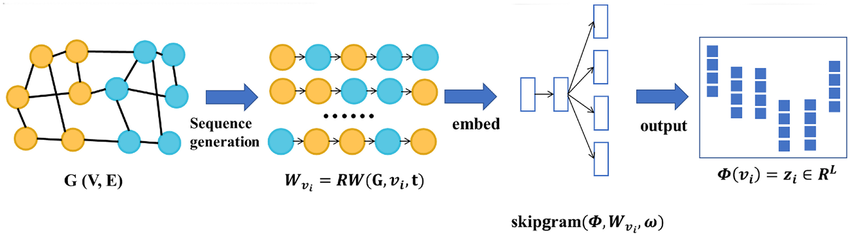
DeepWalk
Definition
DeepWalk is a random walk based Node Embedding method capturing the structural information of a Graph by performing random walks on it. This method can handle large graphs efficiently, and can be applied to various types of graphs (directed, undirected, weighted). However, it does not incorporate node attributes or features.
Algorithm
Link to original
- Run short fixed-length uniform random walks starting from each node in the graph.
- These walks generate sequences of nodes for each node, similar to sentences in natural language processing.
- The sequences are then fed into a Word2Vec-like model to learn vector representations for each node.
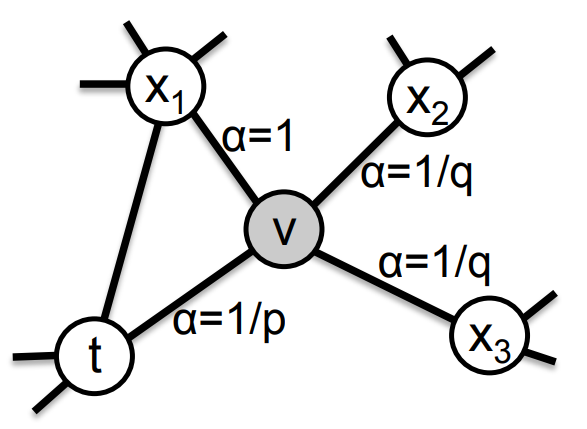
Node2Vec
Definition
Node2Vec is an extension of DeepWalk that introduces more flexible random walks. The model can capture both local and global network structure
Algorithm
- Run short fixed-length biased random walks starting from each node in the graph using some random walk strategy .
Consider a random walk that just traversed edge and now resides at node . The unnormalized transition probability on edges is given by where:
- is the return parameter, controlling the likelihood of immediately revisiting a node in the walk
- is the in-out parameter controlling the search strategy (depth-first vs. breadth-first)
- is the shortest path distance between node and
Link to original
- These walks generate sequences of nodes for each node, similar to sentences in natural language processing.
- The sequences are then fed into a Word2Vec-like model to learn vector representations for each node.
Graph Neural Networks
Graph Neural Network
Definition
Graph neural network (GNN) is a Neural Network model for processing data that can be represented as graphs. The model can learn and extract features from both node attributes and graph structure. The node features are iteratively updated by aggregating information from neighboring nodes.
Link to original
Graph Convolutional Network
Definition
Graph convolutional network (GCN) is a CNN operating on Graph-structured data which takes two inputs: node features and its Adjacency Matrix . GCN learns hidden layer vectors that encode both node features and local graph structures on a fixed graph.
GCN Layer
The GCN layer Is a function of node feature and adjacency matrix. where:
- is the node feature matrix at layer , is the node features.
- is the Adjacency Matrix of the graph
- is the learnable weight matrix for layer It works similar to the filter in CNN with channel size. It is shared across all nodes and independent on the graph size.
- is a non-linear Activation Function
Normalized GCN Layer
To increase numerical stability by enforcing self-loop, the Adjacency Matrix is transformed by adding an identity matrix and normalizing with the Degree Matrix . where
GCN Layer With Self-Transform Term
In some scenario, the self transformation term is added to the GCN layer to help preserve node-specific information where is the learnable self-transformation matrix for layer
GCN Layer With Skip Connection
The skip connection can ease over-smoothing problem.
Neighborhood-Based approach
where is the neighbors of the node .
The aggregation function could be a mean, sum, or max operation:
Facts
The output of a GCN is permutation invariant: invariant to the ordering of nodes in the input graph. The output of each layer in a GCN is permutation equivalent: equivalent to the permutation of nodes in the input graph.
Link to original
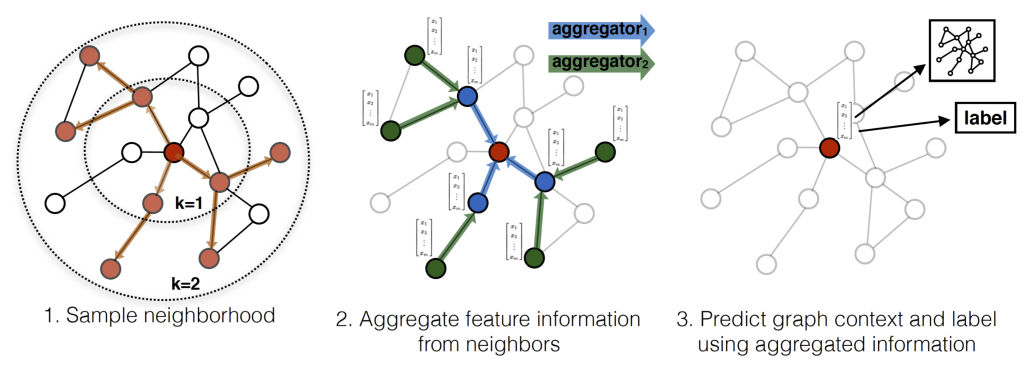
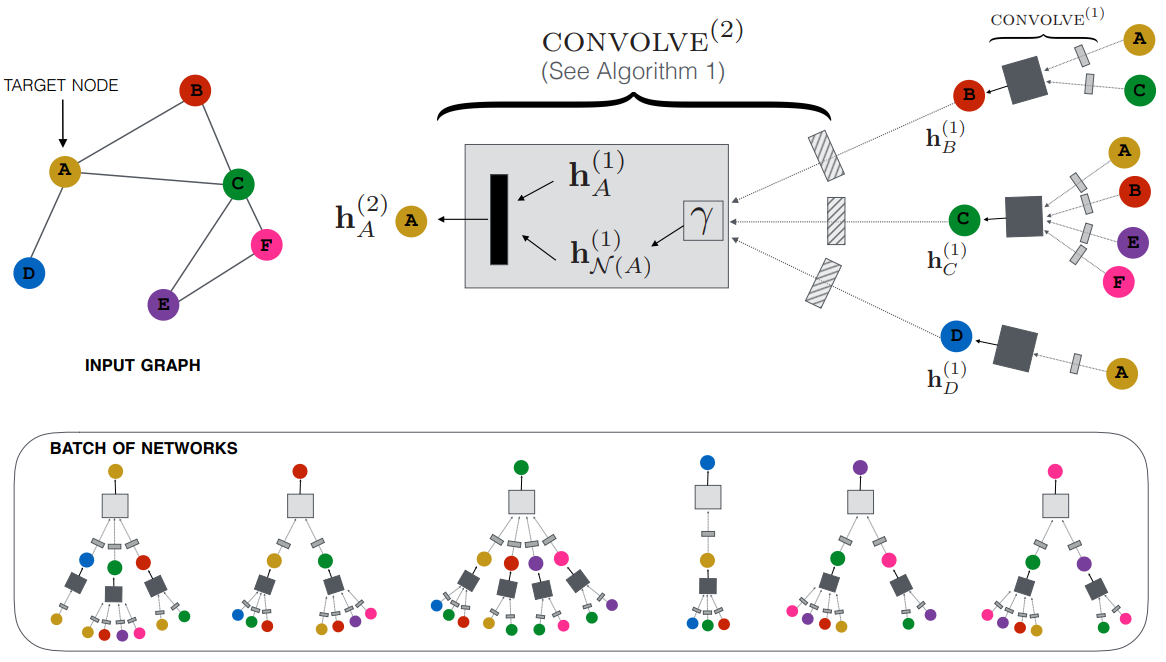
GraphSAGE
Definition
Graph SAmple and aggreGatE (GraphSAGE) is a Node Embedding method sampling and aggregating features from each node’s local neighborhood. It can be generalized to unseen data.
Architecture
GraphSAGE does not require the adjacency matrix of entire graph used in GCN, it utilizes fixed number of neighbors for each node.
Aggregators
Mean aggregator:
LSTM aggregator:
(v_{i})\})$$ where the order of the feature sequence is random Pooling aggregator: $$\operatorname{AGG}_{k}^{\text{pool}} = \max(\{\operatorname{MLP}(h^{(k-1)}_{j})|v_{j}\in \mathcal{N}_{k} (v_{i})\})$$ # Algorithm 1. For $k=1,\dots, K$: 1. For each node $v_{i} \in V$: 1. Sample a fixed-size set of neighbors $\mathcal{N}_{k}(v_{i})$. 2. Aggregate the features of the sampled neighbors using an aggregator function (mean, [[Long Short-Term Memory|LSTM]], pooling). $$h^{(k)}_{\mathcal{N}(v_{i})} = \operatorname{AGG}_{k}(\{h^{(k-1)}_{j}|v_{j}\in \mathcal{N}_{k}(v_{i})\})$$ 3. Combine the aggregated neighborhood information with the node's own features and apply a non-linear [[Activation Function]]. $$h_{i}^{(k)} = \sigma(W^{(k)} \operatorname{CONCAT}(h_{i}^{(k-1)}, h_{\mathcal{N}(v_{i})}^{(k)}))$$ where $\sigma$ is a non-linear [[Activation Function]], and $h_{i}^{(k)}$ is a feature of node $v_{i}$ at $k$-th stage 2. Normalize the feature embedding $$h_{i}^{(k)} = \frac{h_{i}^{(k)}}{||h_{i}^{(k)}||_{2}},\forall v_{i}\in V$$ 2. After $K$ iterations, the final vectors are the output node embeddings. $$z_{i} = h_{i}^{(k)},\forall v_{i} \in V$$Link to original
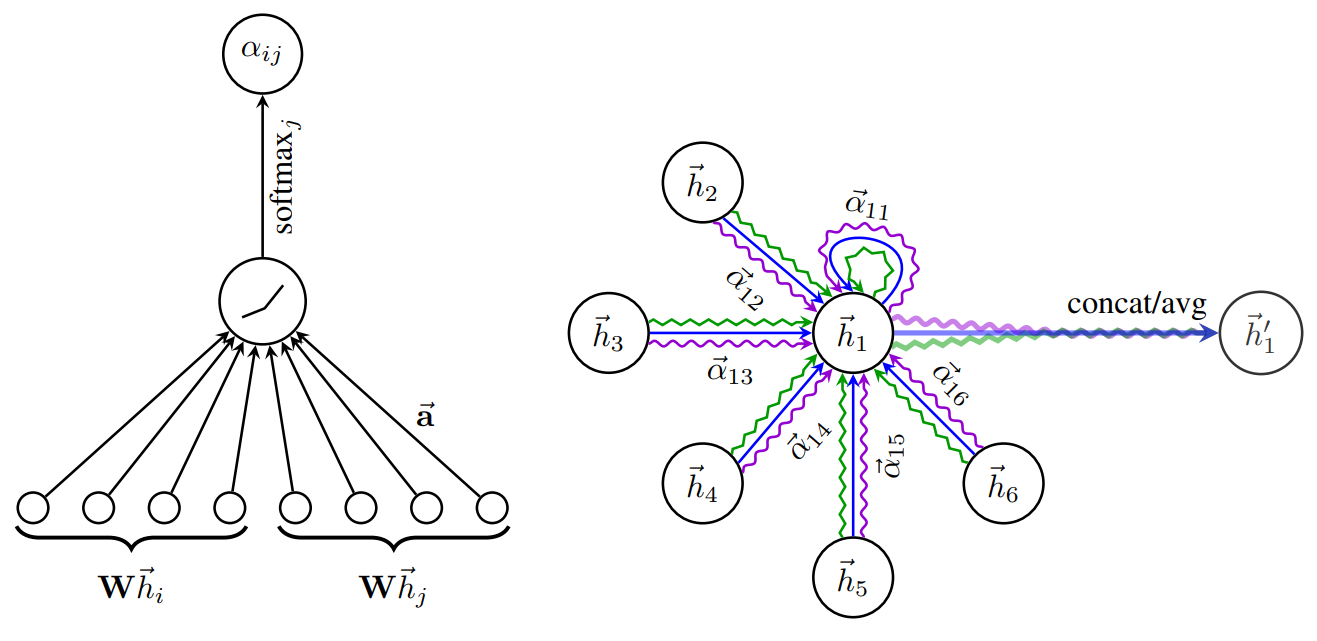
Graph Attention Network
Definition
Graph attention network (GAT) applied Attention mechanism to the aggregation stage of GraphSAGE model. Instead of assigning same weights to all neighbors, GAT uses attention coefficient between nodes.
Architecture
Attention
The attention values are calculated as where and are learnable parameters shared across all nodes.
Aggregation
The feature of node at -th stage is calculated with attention where is a non-linear Activation Function
Multi-Head Attention
To stabilize the learning process, GAT employs multi-head attention. The independent attentions are executed with the same input, and the output of the output features are concatenated or averaged. where , and is the number of attention heads.
Link to original
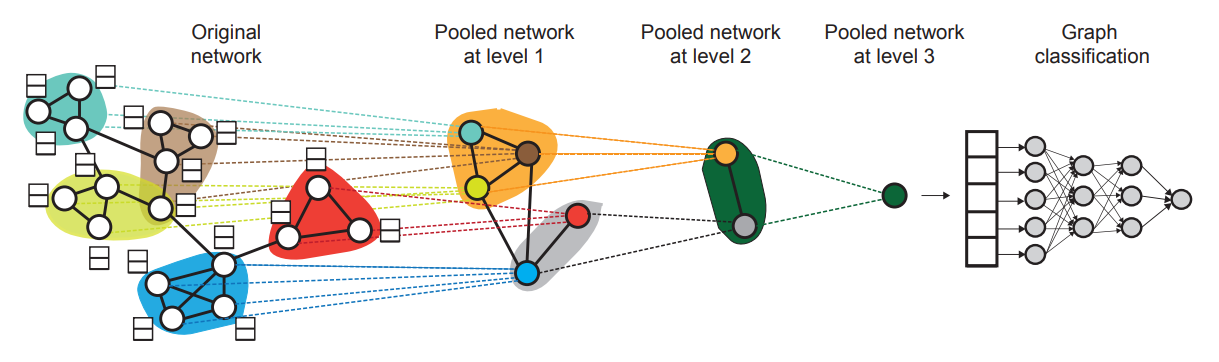
Differentiable Pooling
Definition
Differentiable pooling (DiffPool) is a hierarchical graph pooling method that reduces the size of graph representations. The model is designed for graph-level tasks, such as graph classification, regression, and matching.
Architecture
DiffPool learns a soft assignment matrix that maps nodes in the input graph to a set of clusters and uses the assignment to generate a coarsened graph with fewer nodes.
The DiffPool layer is constructed by the two GNNs: embedding and pooling GNNs.
The embedding GNN is used to generate an embedding matrix The pooling GNN is used to generate an assignment matrix representing a soft clustering of nodes. where and are the input cluster features and adjacency matrices at layer respectively, and the number of columns of is a hyperparameter and smaller than the number of input nodes.
DiffPool layer generates a new coarsened adjacency matrix and a new matrix embedding using the embedding and pooling GNNs.
Link to original
Graph Isomorphism Network
Definition
The Graph isomorphism network (GIN) is a GNN architecture having better expressive power and ability to distinguish different graph structures with injective aggregation.
Architecture
GNNs generates a Node Embedding using the computational graph corresponding to a subtree rooted around each node. The most expressive GNN maps different computational graphs (rooted subtrees) into different node embeddings. If each step of GNN’s neighbor aggregation is injective, then the model retains the full neighboring information. So, the generated node embeddings can distinguish different rooted subtrees.
Any injective multi-set function can be expressed as where the functions and are both some non-linear functions.
Thanks to the Universal Approximation Theorem it can be approximated with multi-layer perceptrons.
GIN uses the injective aggregation with the features of a node and its neighbors.
h_{i}^{(k)} &= \operatorname{MLP}_{\phi}^{(k)}\left((1+\epsilon^{(k)})\operatorname{MLP}^{(k)}_{f}(h_{i}^{(k-1)})+ \sum\limits_{j \in \mathcal{N}(v_{i})}\operatorname{MLP}^{(k)}_{f}(h_{j}^{(k-1)}) \right)\\ &\approx \operatorname{MLP}^{(k)}\left((1+\epsilon^{(k)})h_{i}^{(k-1)} + \sum\limits_{j \in \mathcal{N}(v_{i})} h_{j}^{(k-1)}\right) \end{aligned}$$ where $\epsilon^{k}$ is a learnable scalar ensuring the central node's features.Link to original
Heterogeneous Graphs
Relational GCN
Definition
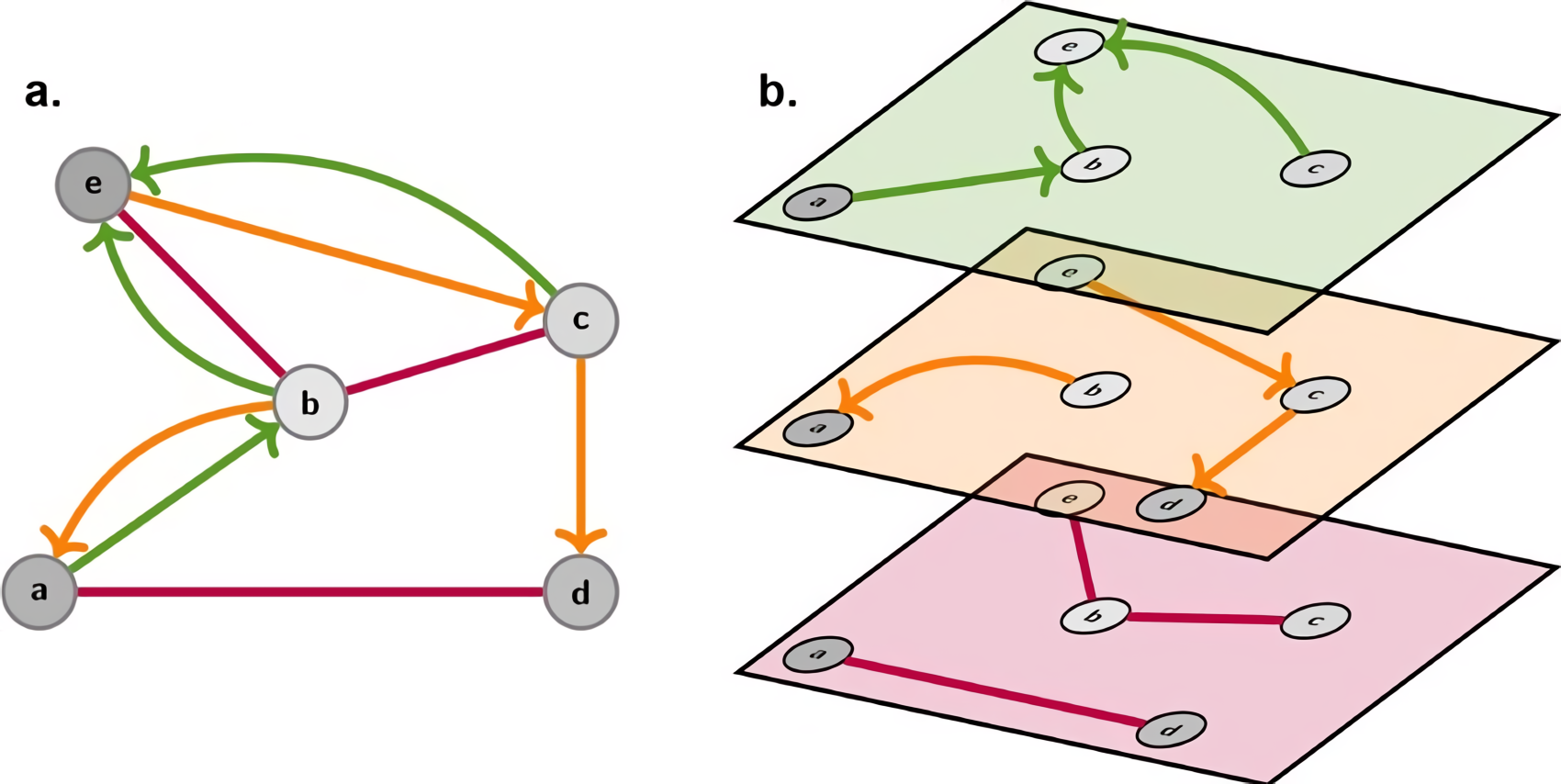
The Relational graph convolutional network (RGCN) is an extension of the GCN designed to handle the graph with multiple types of edges or relationships between nodes.
Architecture
The model uses relation-specific wrights matrices. The information of neighbors of a node is aggregated considering the different relation types.
where:
- is the feature vector of node at layer
- is the set of relations (edge types) in the graph
- is the neighbors of the node under relation .
- is the learnable weight matrix for relation at layer ( is for self-connection).
- is a non-linear Activation Function
Regularization Methods
The number of parameters of the model is increased rapidly with respect to the number of relations. I cause overfitting and scability issues. The techniques such as basis decomposition or block-diagonal decomposition of the weight matrices can be employed to reduce the number of parameters and improve model efficiency.
Basis Decomposition
In basis decomposition, instead of learning a separate weight matrix for each relation, we express each relation-specific weight matrix as a linear combination of a smaller set of basis matrices.
where:
- is the number of basis matrices (typically )
- are the learnable basis matrices
- are learnable importance weight of matrix .
This method reduces the number of parameters from to , where is the hidden dimension in layer .
Block-Diagonal Decomposition
In block-diagonal decomposition, each relation-specific weight matrix is constrained to be block-diagonal
where each is a low-dimensional matrix of size .
This method reduces the number of parameters from to , where is the hidden dimension in layer .
The channels (dimensions) of the node feature vector are indeed grouped The transformation of the channel values occurs within each group
It can be viewed as grouping the channels of the node feature vector and transforming the channel values within each group.
Link to original
Heterogeneous Graph Transformer
Definition
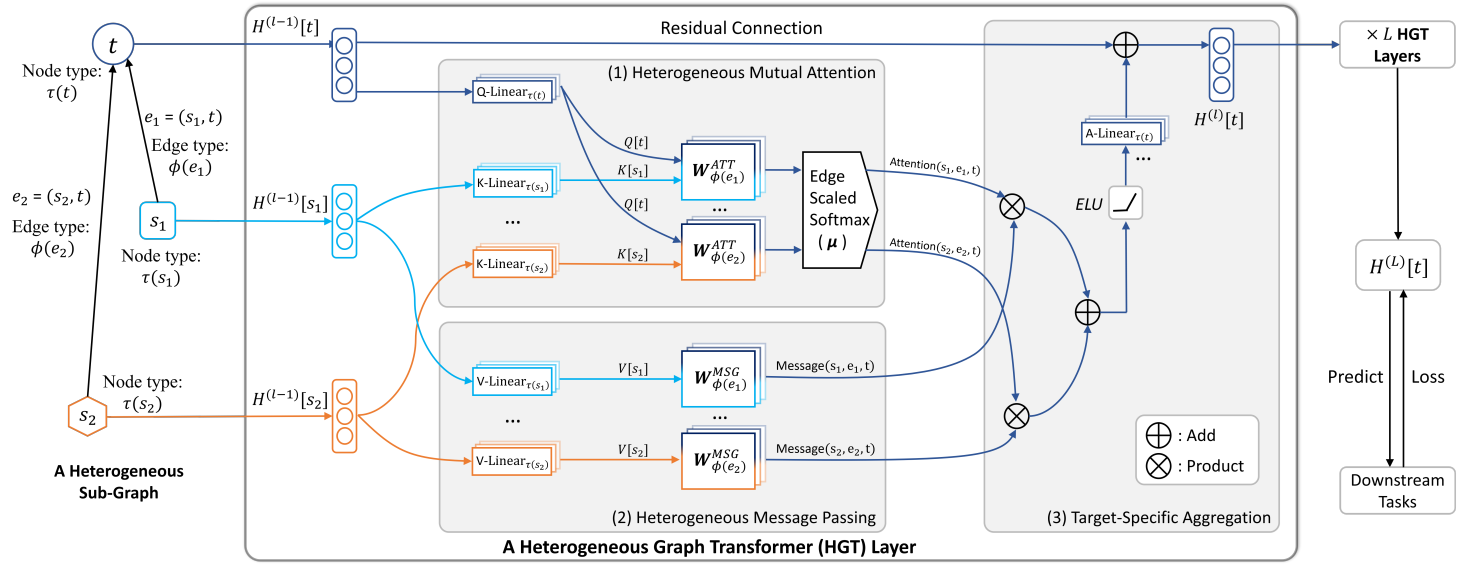
Heterogeneous graph transformer (HGT) is an extension of the GCN designed to handle the graph with multiple types of nodes and edges.
Architecture
Meta-relation triplets
is the target node, and are the source nodes, and are the edges, and the corresponding meta-relations are and .
HGT uses meta-relation triplets (source node type, edge type, target node type) to describe the relationships between different entity types in a graph.
Heterogeneous Mutual Attention
HGT uses an attention mechanism that considers node types and edge types. The attention weights are computed based on the meta-relation triplets, allowing for type-specific information propagation.
For a meta-relation , the attention is calculated by where is the neighbors of the node , and is the number of attention heads.
The attention head is calculated by where:
- is a query vector
- is a key vector
- and are the feature vectors of the source and target nodes at layer .
- , , and are learnable parameter matrices specific to the meta-relation.
Heterogeneous Message Passing
The information of source nodes are passed to the target node. Similar to the attention process, the meta-relations of edges are incorporated into the message passing process.
For a meta-relation , the multi-head message is calculated by where is the number of message heads.
The message head is calculated by where:
- is a value vector
- , are learnable parameter matrices specific to the meta-relation.
Target-Specific Aggregation
The heterogeneous multi-head attention and message are aggregated from the source nodes to the target node. The attention values are used as weights averaging corresponding messages from the source nodes.
The updated vector is calculated by
The final output, contextualized representation of the node, is calculated using the updated vector. where
Link to original
- is the feature vectors of the target node at layer , and works as a residual connection.
- is learnable parameter matrix specific to the meta-relation.
- is a non-linear Activation Function
Knowledge Graphs
Knowledge Graph
Definition
Knowledge graph (KG) is a structured representation of information that organizes data in the form of entities and their relationships. Commonly, knowledge graphs are massive and incomplete. So, knowledge graph completion (KG completion) is an important task.
Knowledge Graph Completion Task
Edges in KG are represented as triplets , where is a head, is a relation, and is a tail. Given a triplet , the goal of KG completion is that the embedding of should be close to the embedding of . A score function is high if input triplet is probable, otherwise low.
Relation Patterns in Knowledge Graph
Types of relations embedding models can represent
Predictive Queries on Knowledge Graph
Examples
Link to original
Node types: drug, disease, adverse event, protein, pathways Relation types: has_func, causes, assoc, treats, is_a
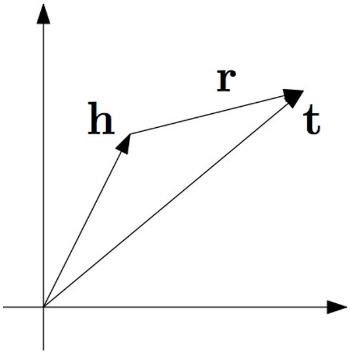
TransE
Definition
Translating embeddings (TransE) is a Knowledge Graph embedding method. TransE represents entities and relations as vectors in the same low-dimensional space. The model is trained through Contrastive Learning.
For a triplet , let be embedding vectors. The objective of TransE is finding the vectors satisfying the equality
The entity score function is defined as
TransE cannot model symmetric and 1-to-N relations
Link to original
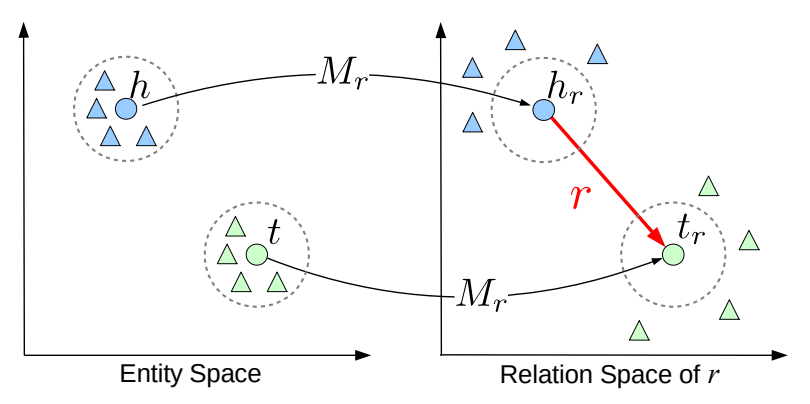
TransR
Definition
Translating embeddings in relational space (TransR) is a Knowledge Graph embedding method. it extends the ieda of TransE model by introducing separate embedding space for entities and relations.
For a triplet , let be vectors in the entity space, and be a vector in relation space.
The objective of TransR is finding the vectors satisfying the equality where and are the projections of and in the relation space, and is the projection matrix of .
The entity score function is defined as
Link to original
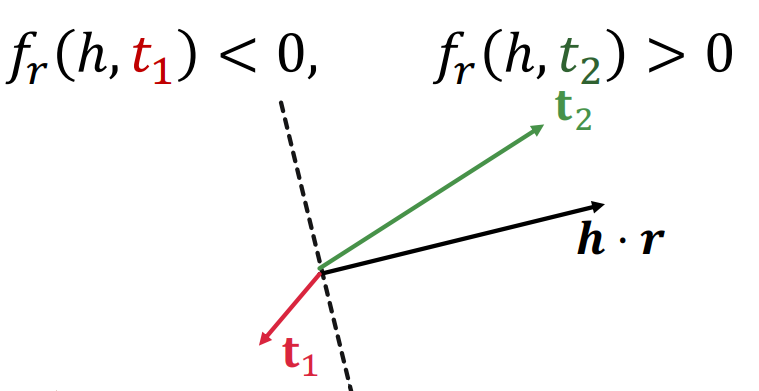
DistMult
Definition
DistMult is a Knowledge Graph embedding method representing entities and relations as vectors in same low-dimensional space. It uses a bilinear scoring function with a diagonal matrix for each relation.
For a triplet , let be embedding vectors.
The score function of DistMult is defined as
DistMult cannot model antisymmetric, inverse, and transitive relations.
Link to original
ComplEx
Definition
Complex embedding (ComplEx) is a Knowledge Graph embedding method. It extends the idea of DistMult in a vector space to the complex domain.
For a triplet , let be complex embedding vectors.
The score function of ComplEx is defined as
ComplEx cannot model transitive relations
Link to original
Reasoning over Knowledge Graphs
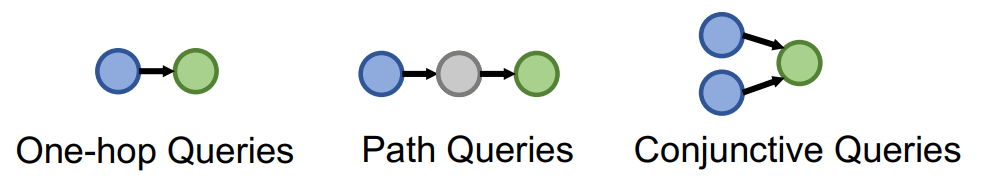
One-Hop Query
Definition
One-hop query is the same task as knowledge graph completion task. KG completion: Is link in the KG? One-hop query: Is an answer to query ?
Link to original
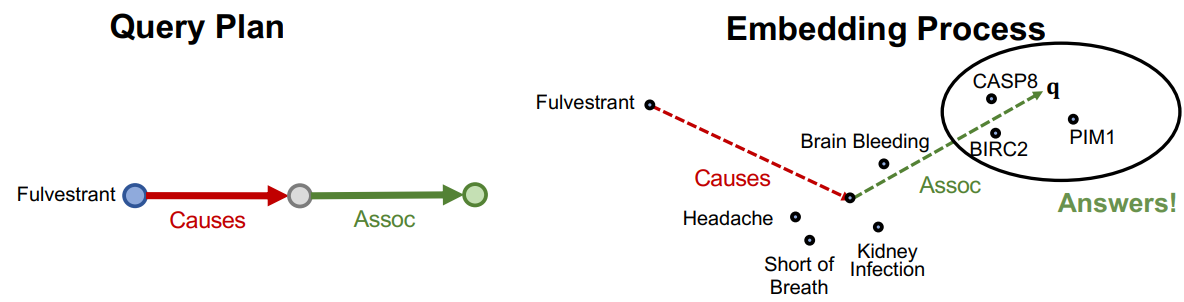
Path Query
Definition
Path query is the generalization of One-Hop Query with more relations on the path.
An -hop path query can be represented by where is an anchor entity, and is an -th relation.
Traversing Knowledge Graph
KGs are incomplete, so we cannot identify all the answer entities. For this reason, the path-based queries over an incomplete Knowledge Graph are calculated by the KG embedding method that can handle transitive relations such as TransE.
Examples
Link to original
Query: (Fulvestrant, (Causes, Assoc))
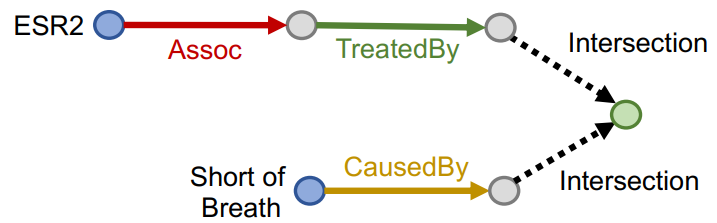
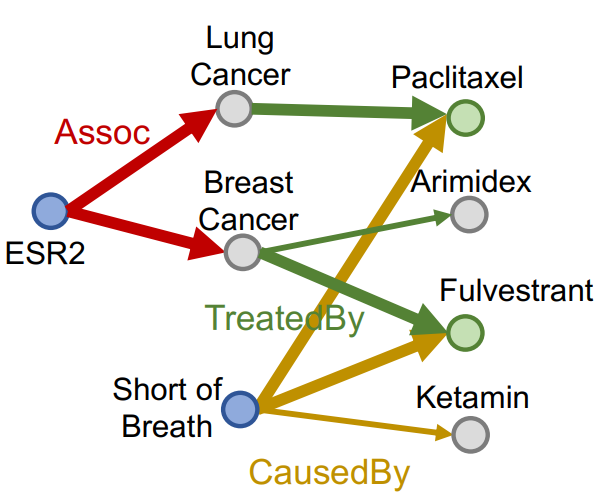
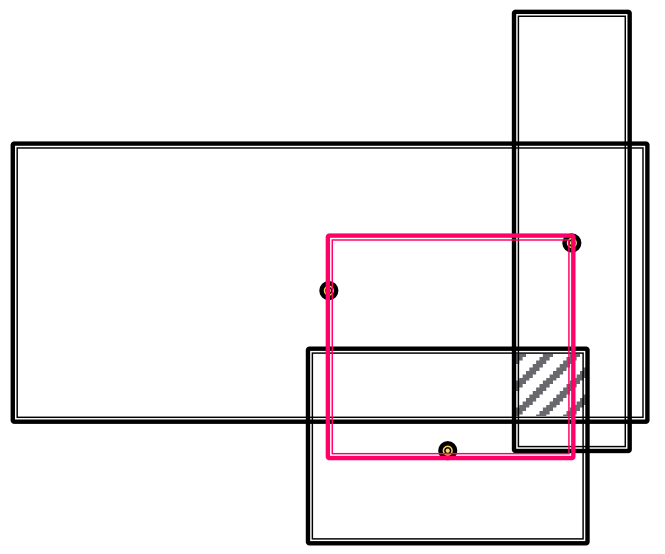
Conjunctive Query
Definition
Conjunctive query has multiple anchor entities having their own path queries.
A conjunctive query can be represented by where is an anchor entity of -th Path Query, and is a -th relation of -th path.
Traversing Knowledge Graph
Each intermediate node of a conjunctive query represents a set of entities, so we can not use the method used for Path Query. The Query2Box model can handle the problem.
Link to original
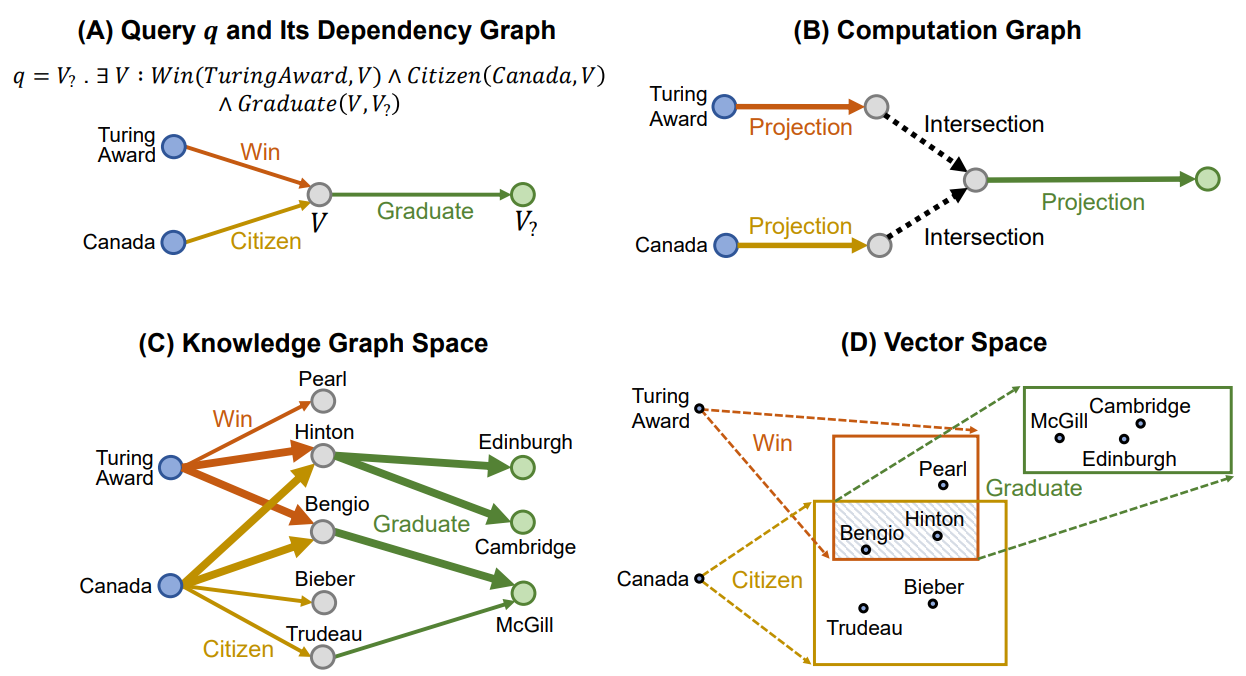
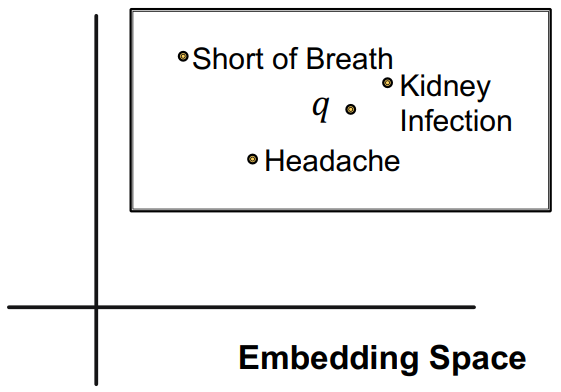
Query2Box
Definition
Query2Box is a model for calculating path-based queries over an incomplete Knowledge Graph. It represents queries as boxes in a vector space. To answer a query, the model computes the final query box and ranks entities based on their distance to the box.
Architecture
Box Embedding
In the Query2Box model, queries are represented as boxes in a vector space, and entities are represented as points (zero-size box) in the space. The box embedding allows for modeling uncertainty and capturing a set of possible answers.
Query box in is defined as where is the center of the box, and is the offset of the box.
The embedding box is defined using the query box. where is element-wise inequality.
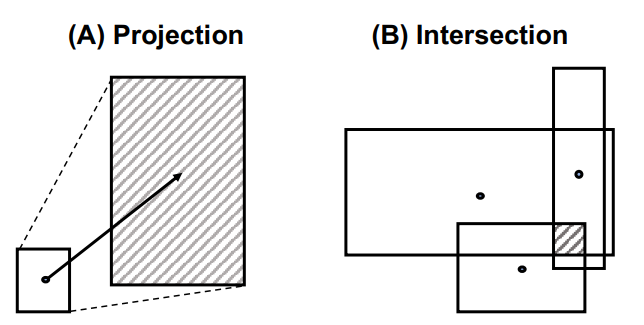
Query Operations
Query2Box consists of the two operations: projection, and intersection.
Projection Operation
The projection operation expands the box to include related entities.
Intersection Operation
The intersection operation combines information from multiple sub-queries by intersecting their boxes. Instead of directly applying intersection over a set of box embeddings, the intersection in calculated using the Attention-like operation.
The center is calculated as where
The offset is calculated as where is a Sigmoid Function, and
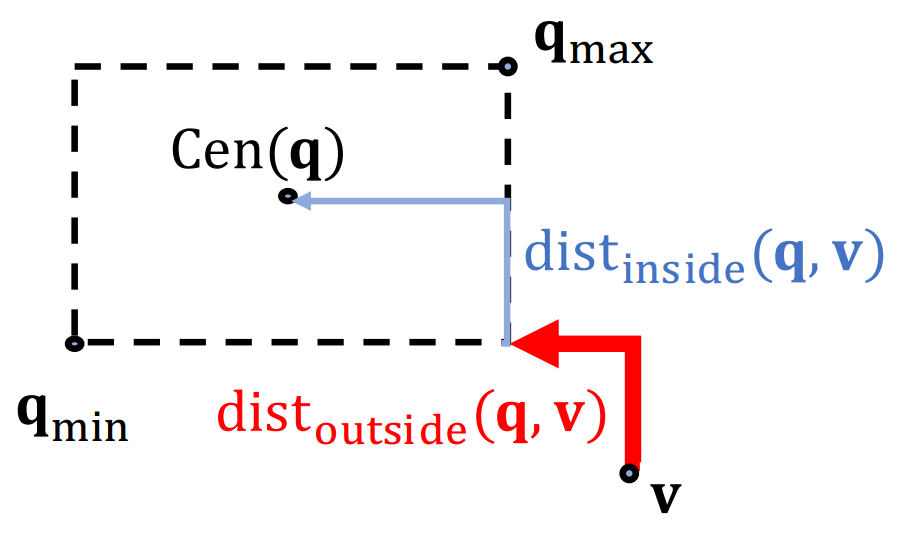
Entity-to-Box Distance
Given a query box and an entity vector , the distance between them is defined as where is a fixed scalar that downweights the distance inside the box.
Link to original
Sugraph and Network Motifs
Subgraph
Definition
A subgraph of a Graph is another graph formed from a subset of the vertices and edges of .
Node-Induced Subgraph
Take subset of the nodes and all edges induced by the nodes.
where and .
Edge-Induced Subgraph
Take subset of the edges and all corresponding nodes. where and .
Link to original
Graph Isomorphism
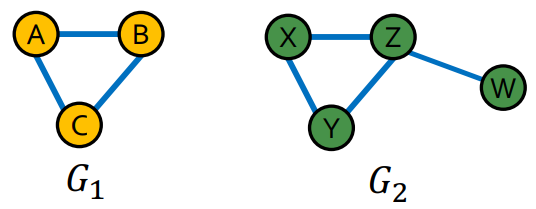
Definition
An isomorphism of Graph and is a Bijection between the vertex sets of the graphs such that any two vertices and of are adjacent in if and only if and are adjacent in .
Is there exist an isomorphism between two graphs, the graphs are isomorphic.
Link to original
Subgraph Isomorphism
Definition
Given two graphs and , if contains a Subgraph that is isomorphic to . where means Graph Isomorphic.
Then is subgraph isomorphic to
Link to original
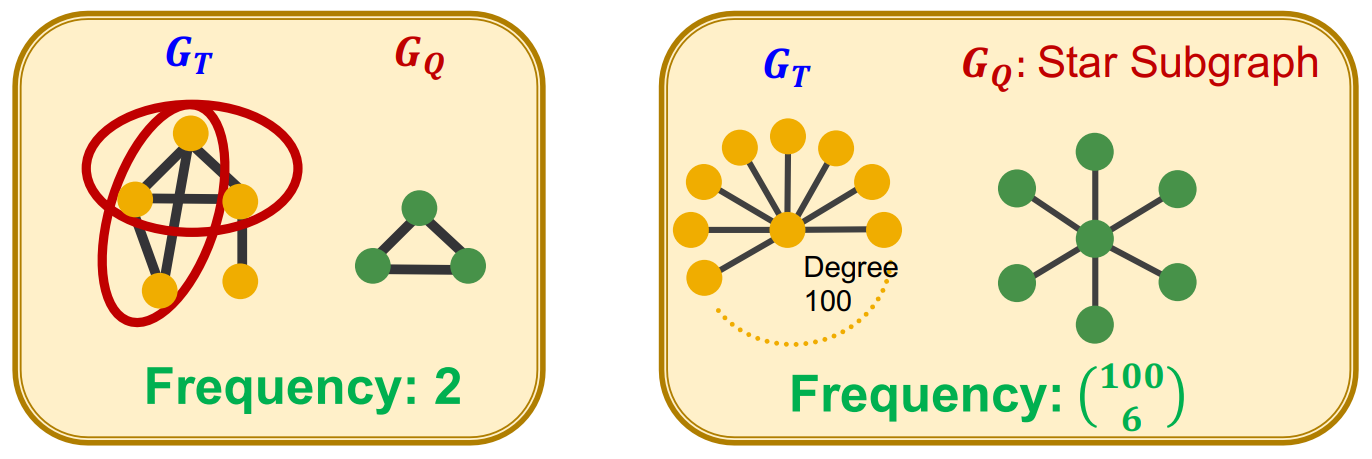
Graph-Level Subgraph Frequency
Definition
Graph-level subgraph frequency refers to the number of times a particular Subgraph pattern appears in the entire network.
Given a Graph and a Subgraph of interest , the graph-level frequency of in is calculated as where means Graph Isomorphic.
Link to original
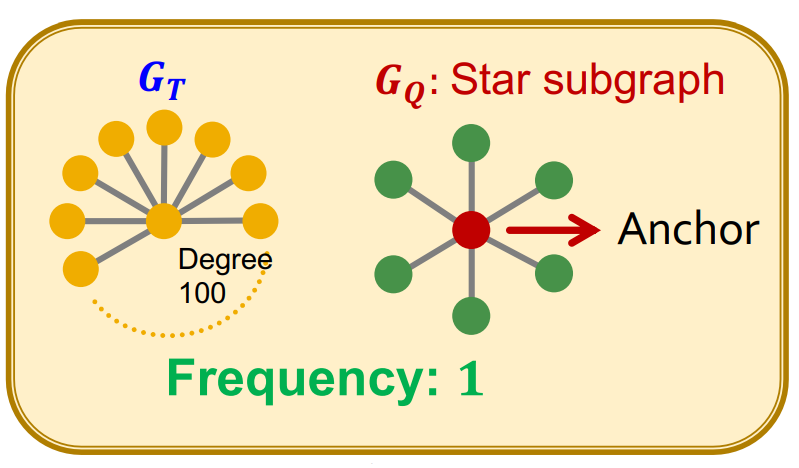
Node-Level Subgraph Frequency
Definition
Given a Subgraph of interest , its node (the anchor), and a Graph , the node-level frequency is defined as the number of nodes for which some Subgraph is isomorphic to a specific Subgraph pattern and the isomorphism maps node to .
where means isomorphic, is an isomorphism, and is the set of vertices in Graph .
Link to original
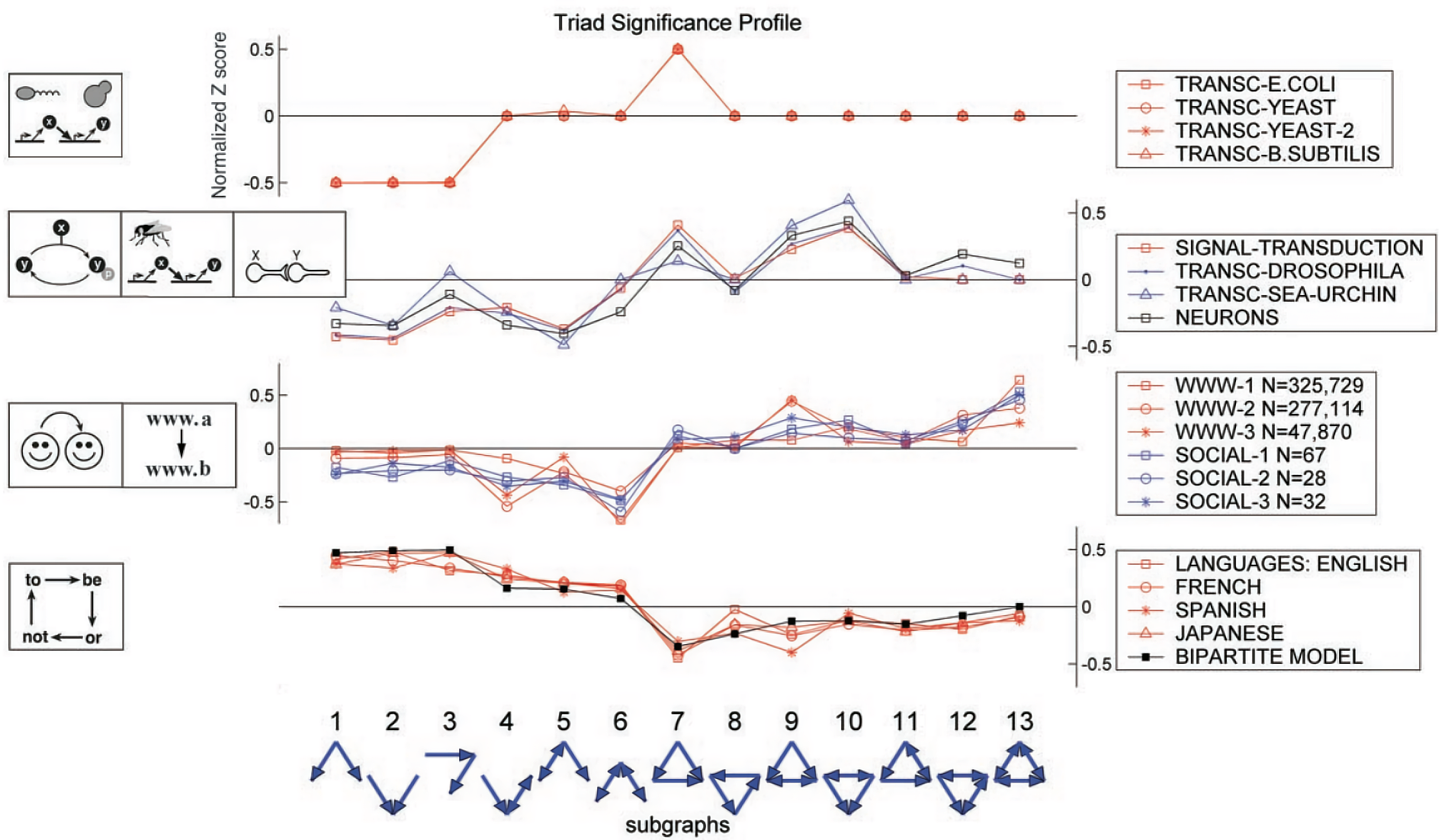
Network Motif
Definition
Network motifs are recurrent and significant interconnected subgraphs of a larger Graph. To be considered a motif, a sub graph must occur more frequently than expected by chance in randomized graphs with same statistics (e.g. number of nodes, edges, degree sequence).
Z-Score
The significance of a network motif can be measured by the Z-score. where is occurrence of the subgraph in , is the average of occurrence of the subgraph in , and is the standard deviation of occurrence of the subgraph in .
The occurrences are measured by Graph-Level Subgraph Frequency or Node-Level Subgraph Frequency.
A high Z-score indicates that the Subgraph appears significantly more often in the real network than expected by chance, suggesting it may be a functionally important motif.
Network Significance Profile
Motifs performs specific functions within a graph, contributing to the overall behavior of it. Different types of graphs (e.g. gene regulation networks, neural networks, social networks, and word connectivity) often have characteristic motifs.
Link to original
Subgraph Matching
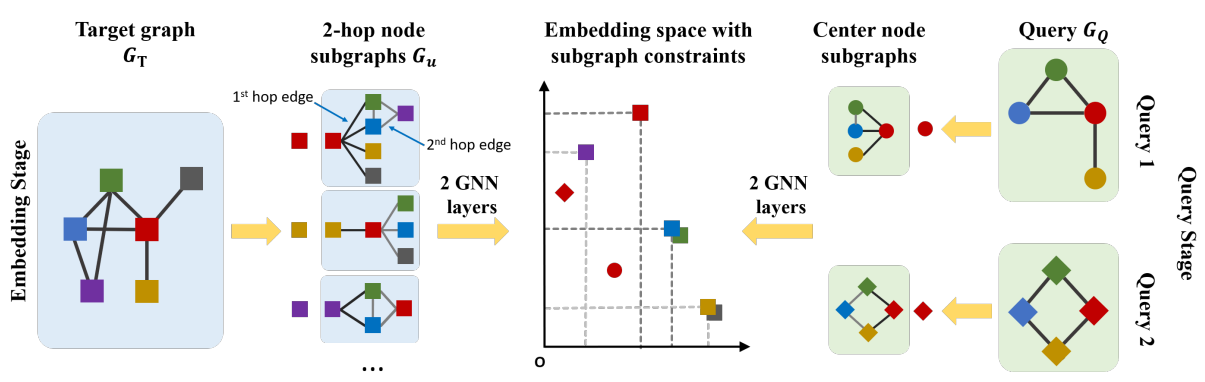
Neural Subgraph Matching
Definition
Neural subgraph matching is a model desinged to solve the subgraph matching problem using neural networks.
Architecture
Consider a subgraph and a target graph
Embedding
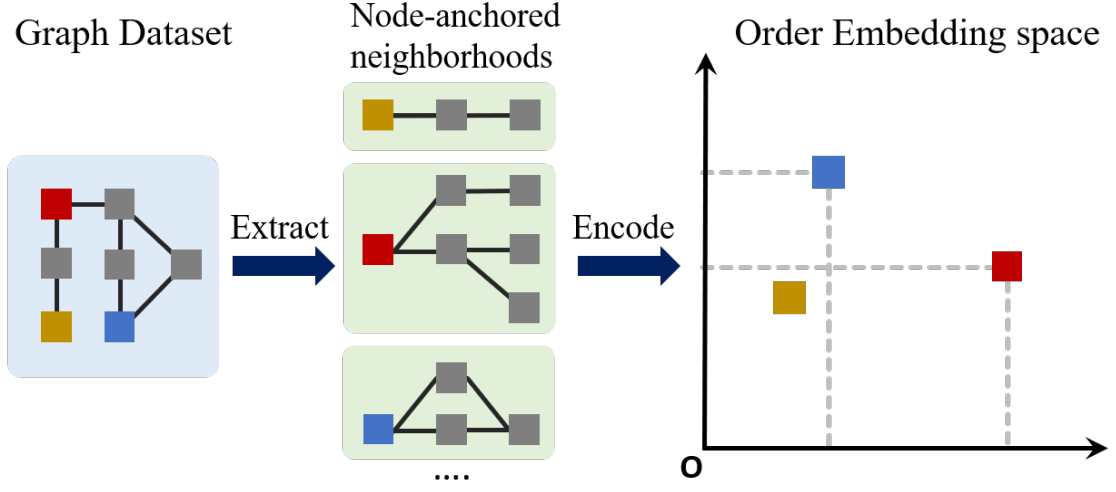
For each node in the query and target graph , obtain a -hop neighborhood around the anchor. The neighborhoods are embedded using GNN by computing the embeddings for the anchor nodes in their respective neighborhoods. The GNN is trained to have partial order relation in its embedding space.
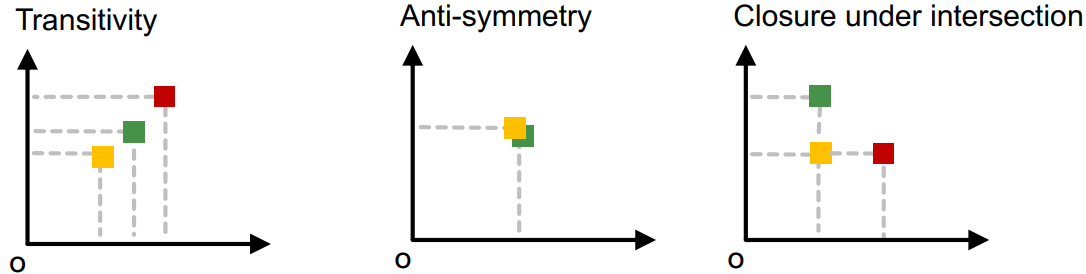
Order Embedding
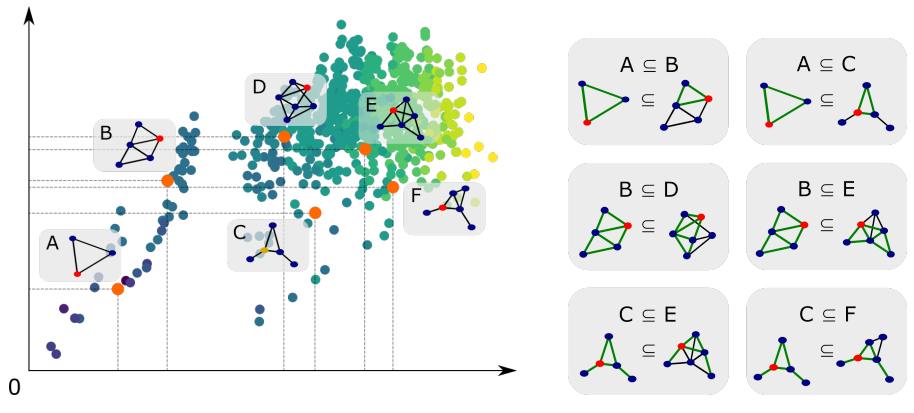
Order embedding enforces a partial order relationship between node embeddings of the sub graphs. It is used to capture the hierarchical relationship between graphs and subgraphs. A partial order is defined in the embedding space where larger graphs are located above of their subgraphs.
Subgraph isomorphism relationship can be encoded in order embedding space where is the dimension of the embedding space, and is the -th value of the embedding vector of subgraph anchor .
The GNN for the embedding is trained by minimizing a max-margin loss with Contrastive Learning examples. The margin between graph embedding of and are defined as The max-margin loss is defined as where is a set of positive pairs, and is a set of negative pairs.
Max-margin loss prevents the model from embedding all vectors to far from the origin.
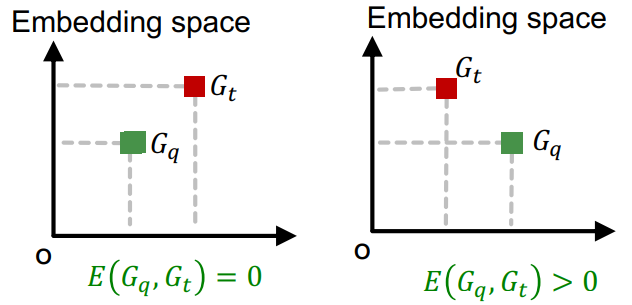
Subgraph Prediction
Consider a query graph anchored at node , and a target graph anchored at node . By using the margin used for embedding, we can check whether is a node-anchored subgraph of the target graph . where is a hyperparameter works as a threshold.
To check if is isomorphic to a subgraph of , repeat this process for all and and aggregate the result to make the binary prediction for the decision problem of subgraph matching. Here is the neighborhood around the anchor node .
Link to original
SPMiner
Definition
Subgraph pattern miner (SPMiner) is a model for identifying frequent subgraphs (network motifs) in a graph.
Architecture
The frequent subsampling counting problem consists of two stages: searching over all possible motifs, and counting the frequency of each motif in graph. SPMiner consists of two steps: Embedding candidate subgraphs, and motif search procedure.
Embedding candidate subgraphs
SPMiner decomposes the input Graph into many overlapping node-anchored neighborhoods around each node. Then encodes each subgraph into an order embedding space.
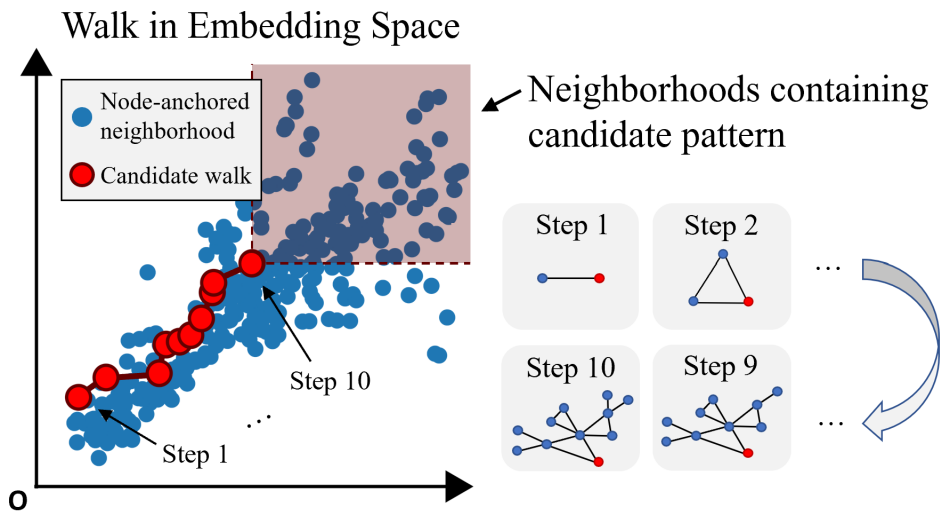
Motif Search Procedure
SPMiner then directly reasons in the embedding space to identify frequent motifs. It searches for a -step walk in the embedding space that stays to the lower left of as many neighborhoods as possible. The walk is performed by iteratively adding nodes and edges to the current motif candidate, and tracking its embedding.
Given an order embedding encoder , let a Graph generation procedure be , where at any step , is generated by adding node to . Then the sequence of embeddings is a monotonic walk in the order embedding space. Finding the frequent motif is the same as finding a walk that
SPMiner can quickly count the number of occurrences of a given motif by simply checking the number of neighborhoods that are embedded to the top-right of it in the embedding space
Link to original
GNNs for Recommendation System
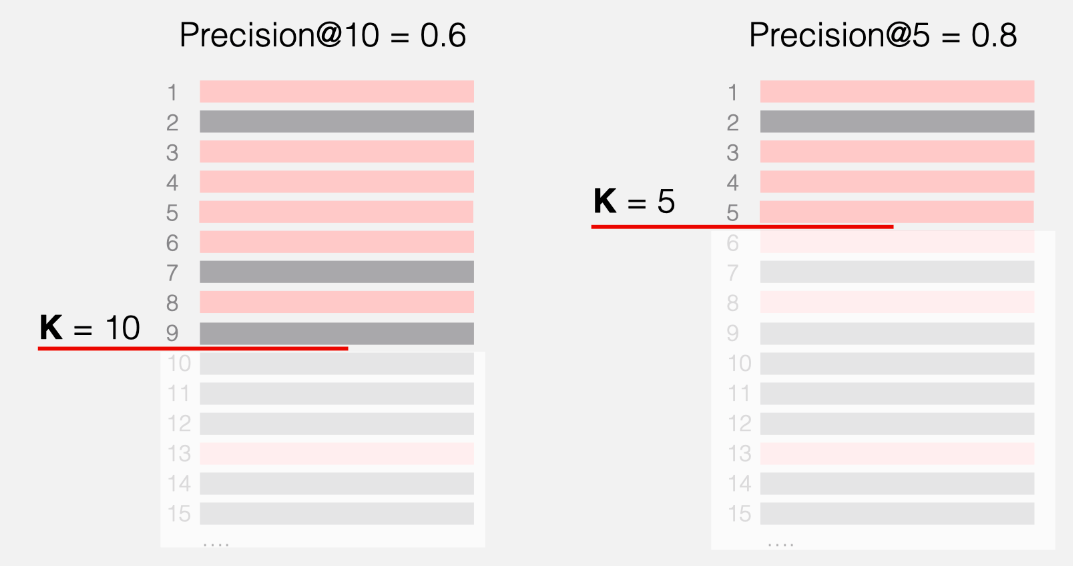
Recall at K
Definition
Recall at K (Recall@K) is a metric that help evaluate the performance of recommendation system. It is the proportion of correctly identified relevant items in the top K recommendations out of the total number of relevant items in the dataset.
For each user , let be a set of positive items the user will interact, and be a set of items recommended by the model. In top-K recommendation .
Recall@K is not differentiable.
Link to original
Binary Loss
Definition
Binary loss is a metric that help evaluate the performance of recommendation system. It treats the recommendation problem as a binary classification task.
Let be a set of all users, be a set of all items, be a set of observed user-item interactions, and be a set of negative edges. The binary loss function is defined as where:
- is a Sigmoid Function
- and are the embedding vectors of and .
- is the score between and .
Since the binary loss is non-personalized, the all positive edges are pushed higher than those of all negative edges.
Link to original
Bayesian Personalized Ranking
Definition
Bayesian personalized ranking (BPR) is a metric that help evaluate the performance of recommendation system. It focuses on the relative order of items rather than absolute scores.
Let be a set of all users, be a set of all items, be a set of observed user-item interactions, and be a set of negative edges.
For each user , the rooted positive and negative edges are defined as The BPR loss for user is defined as
The final BPR loss is the average of them. where:
Link to original
- is a Sigmoid Function
- and are the embedding vectors of and .
- is the score between and .
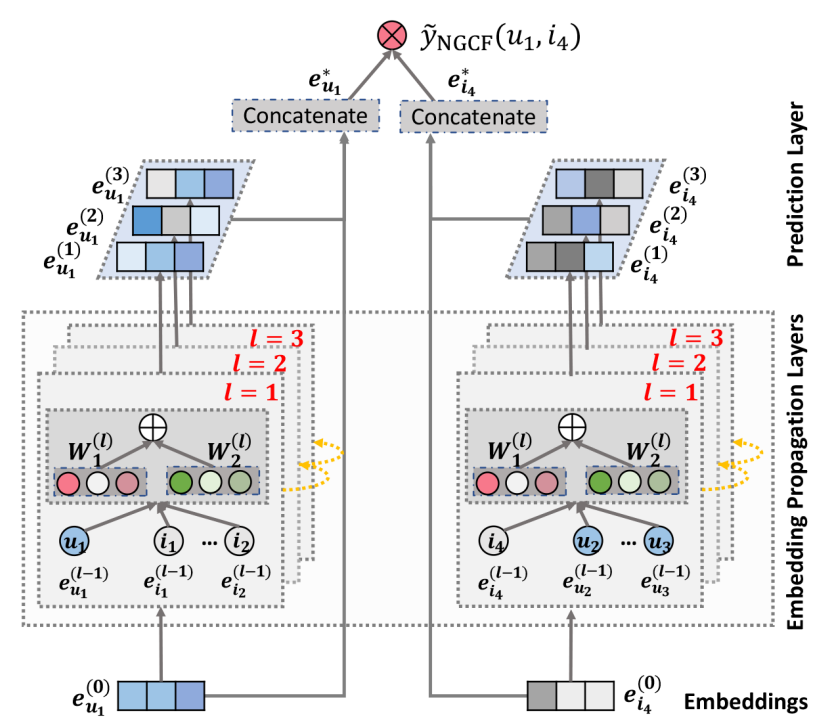
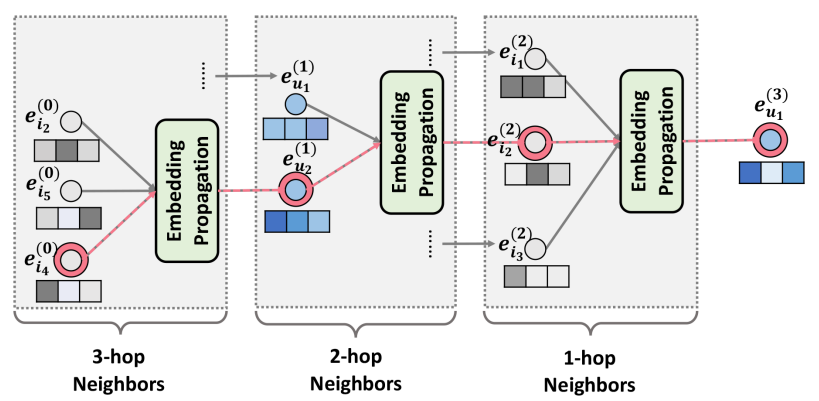
Neural Graph Collaborative Filtering
Definition
Neural graph collaborative filtering (NGCF) is a recommendation algorithm that uses GNN to enhance collaborative filtering.
Architecture
NGCF constructs a bipartite graph where users and items are nodes, and interactions form edges.
Embedding Propagation Layers
The embeddings of NGCF are updated through propagation.
\mathbf{e}_{u}^{(l+1)} &= \sigma\left( \sum_{i \in \mathcal{N}_{u} \cup \{u\}} \frac{1}{\sqrt{|\mathcal{N}_{u}||\mathcal{N}_{i}|}} (W_{1}\mathbf{e}_{i}^{(l)} + W_{2}(\mathbf{e}_{i}^{(l)} \odot \mathbf{e}_{u}^{(l)})) \right)\\ \mathbf{e}_{i}^{(l+1)} &= \sigma\left( \sum_{i \in \mathcal{N}_{i} \cup \{i\}} \frac{1}{\sqrt{|\mathcal{N}_{u}||\mathcal{N}_{i}|}} (W_{1}\mathbf{e}_{u}^{(l)} + W_{2}(\mathbf{e}_{i}^{(l)} \odot \mathbf{e}_{u}^{(l)})) \right) \end{aligned}$$ where - $\mathbf{e}_{u}^{(l)}$ is the embedding of user $u$ at the $l$-th layer - $\mathbf{e}_{i}^{(l)}$ is the embedding of item $i$ at the $l$-th layer - $\mathcal{N}_{u}$ is the set of items interacted with by user $u$. - $W_{1}$ and $W_{2}$ are learnable weight matrices - $\odot$ denotes element-wise multiplication - $\sigma$ is a non-linear [[Activation Function]] It can be written in a matrix form. $$E^{(l+1)} = \sigma\left( (\mathcal{L}+I) E^{(l)} W_{1}^{(l+1)} + \mathcal{L} (E^{(l)} \odot E^{(l)}) W_{2}^{(l+1)} \right)$$ where: - $\mathcal{L} = D^{-\frac{1}{2}}AD^{-\frac{1}{2}}$ is the [[Laplacian Matrix]] for the user-item graph - $A = \begin{bmatrix} O&R\\R^{\intercal}&O \end{bmatrix}$ is the [[Adjacency Matrix]], where $R$ is $N \times M$ user-item interaction matrix - $D$ is the [[Degree Matrix]] - $W_{1}^{(l+1)}$ and $W_{2}^{(l+1)}$ are learnable weight matrices at layer $k+1$ ## Model Prediction The final user and item embedding matrices are constructed by concatenating all the layer's embeddings $$\begin{aligned}\mathbf{e}_{u}^{*} &= \operatorname{CONCAT}(\{\mathbf{e}_{u}^{(l)}|l=1,\dots,L\})\\ \mathbf{e}_{i}^{*} &= \operatorname{CONCAT}(\{\mathbf{e}_{i}^{(l)}|l=1,\dots,L\}) \end{aligned}$$ where $L$ is the total number of layers. The score between $u$ and $i$ is calculated using the inner product $$\operatorname{score}_{\text{NGCF}}(u, i) = {\mathbf{e}_{u}^{*}}^{\intercal}\mathbf{e}_{i}^{*}$$Link to original
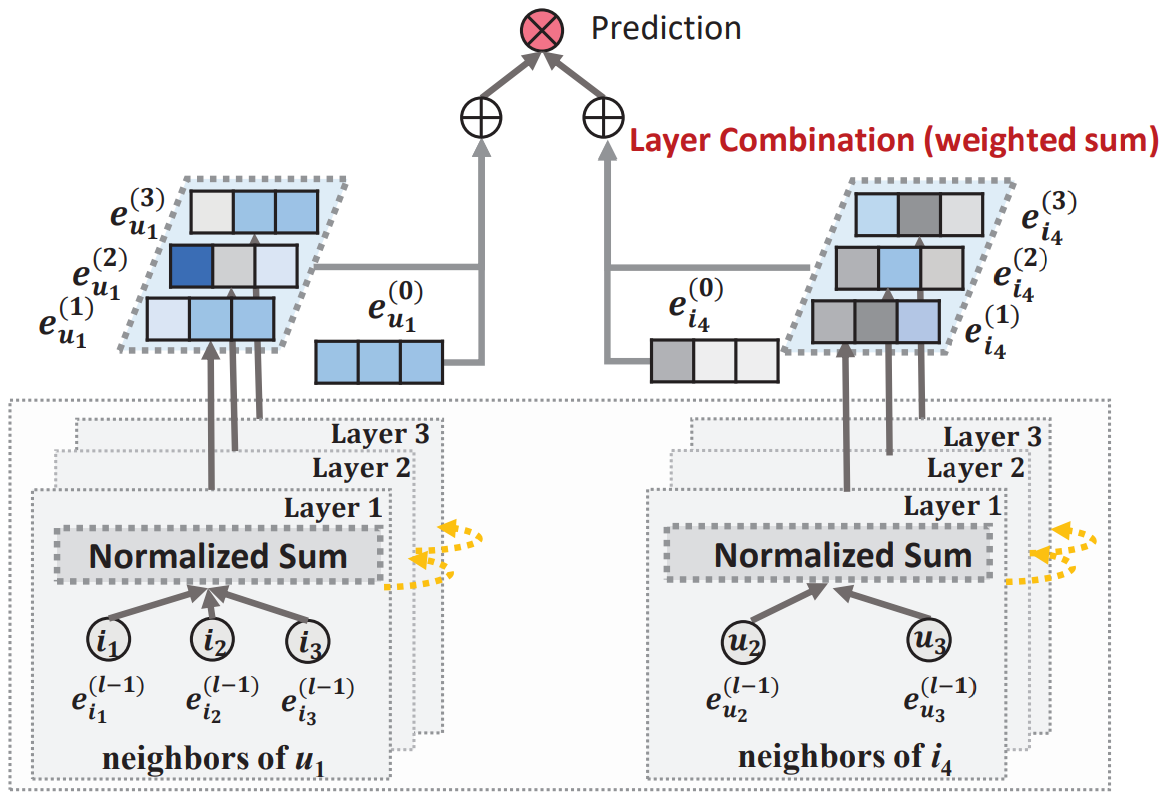
LightGCN
Definition
LightGCN (Light Graph Convolutional Network) is a simplified version of NGCF.
Architecture
Light Graph Convolution
In the LGC, only the normalized sum of neighbor embeddings is performed towards next layer; other operations like self-connection, feature transformation, and nonlinear activation are all removed.
The graph convolution operation in LightGCN is defined as
\mathbf{e}_{u}^{(l+1)} &= \sum_{i \in \mathcal{N}_{u}} \frac{1}{\sqrt{|\mathcal{N}_{u}||\mathcal{N}_{i}|}} \mathbf{e}_{i}^{(l)}\\ \mathbf{e}_{i}^{(l+1)} &= \sum_{u \in \mathcal{N}_{i}} \frac{1}{\sqrt{|\mathcal{N}_{u}||\mathcal{N}_{i}|}} \mathbf{e}_{u}^{(l)} \end{aligned}$$ where - $\mathbf{e}_{u}^{(l)}$ is the embedding of user $u$ at the $l$-th layer - $\mathbf{e}_{i}^{(l)}$ is the embedding of item $i$ at the $l$-th layer - $\mathcal{N}_{u}$ is the set of items interacted with by user $u$. It can be written in a matrix form. $$E^{(l+1)} = \mathcal{L} E^{(l)}$$ where: - $\mathcal{L} = D^{-\frac{1}{2}}AD^{-\frac{1}{2}}$ is the normalized adjacency matrix for the user-item graph - $A = \begin{bmatrix} O&R\\R^{\intercal}&O \end{bmatrix}$ is the [[Adjacency Matrix]], where $R$ is $N \times M$ user-item interaction matrix - $D$ is the [[Degree Matrix]] The only learnable parameters are the embeddings at the $0$-th layer. ## Model Prediction The final user and item embedding matrices are constructed by the weighted sum of the embeddings in each layer. $$E_{\text{final}} = \alpha_{0}E^{(0)} + \alpha_{1}E^{(1)} + \dots + \alpha_{K}E^{(K)}$$ where $\alpha_{k}$ is a hyperparameter (The parameters are set uniformly $\alpha_{k} = \cfrac{1}{K+1}$ in the paper). The score between $u$ and $i$ is calculated using the inner product $$\operatorname{score}_{\text{LGC}}(u, i) = {\mathbf{e}_{u}^{*}}^{\intercal}\mathbf{e}_{i}^{*}$$Link to original
PinSAGE
Definition
PinSAGE (Pinterest SAGE) is a Graph Convolutional Network model for large-scale recommendation systems.
Architecture
PinSAGE is based on the GraphSAGE architecture.
Importance-Based Neighbors
We simulate random walks starting from node and count the visits for each node in the random walk. The neighborhood of , , is then defined as the top- nodes with the highest visit counts with respect to node .
Importance Pooling
PinSAGE assigns different weights (normalized visit counts) to different neighbors when aggregating information from a node’s neighborhood. This allows the model to focus on more relevant or influential neighbors.
Curriculum Training
PinSAGE implements a curriculum training strategy, starting with easier examples and gradually moving to more difficult ones. The difficulty of negative samples is based on the visit counts
Link to original
Deep Generative Models for Graphs
GraphRNN
Definition
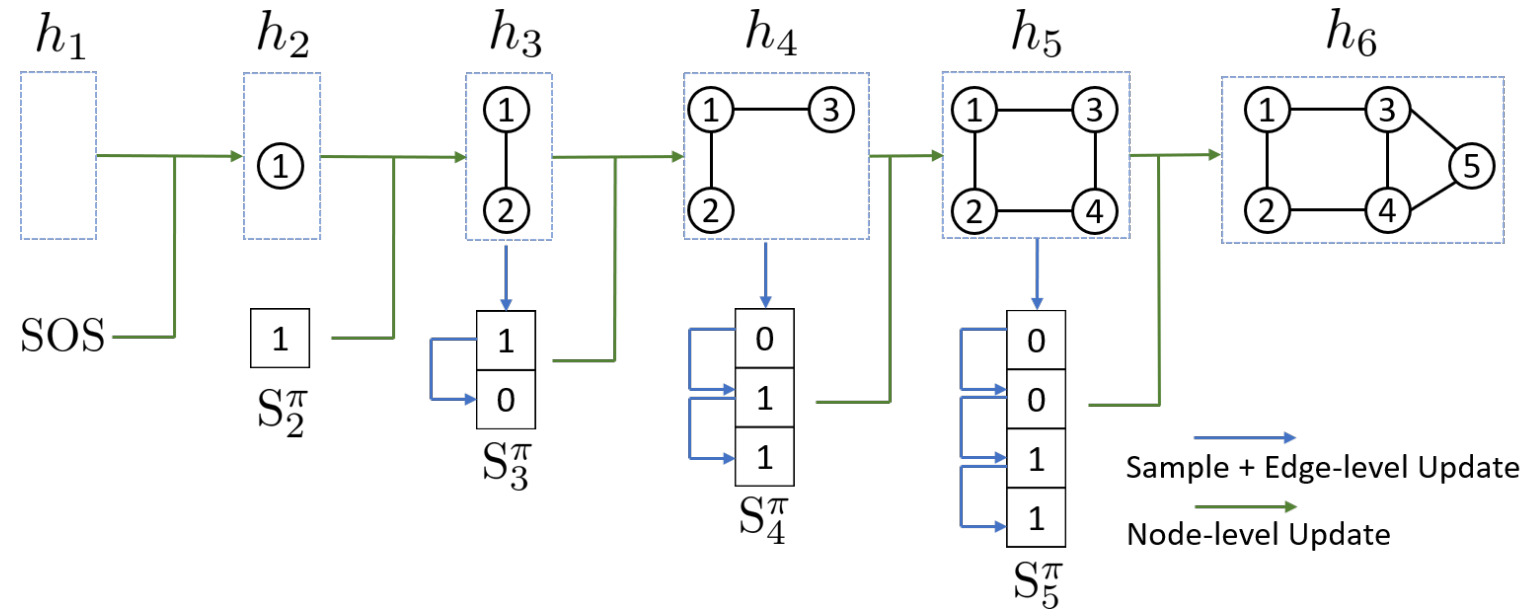
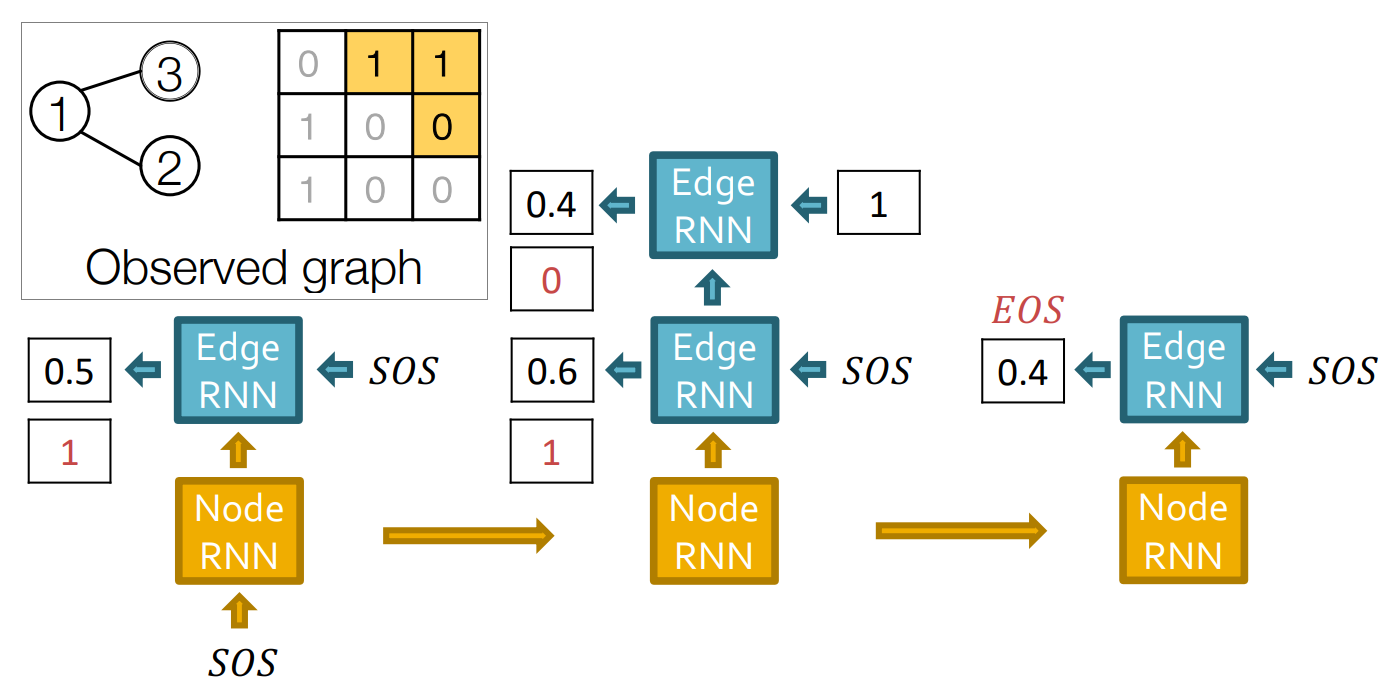
Graph recurrent neural network (GraphRNN) is a graph generation model. It generates graph in a autoregressive manner. GraphRNN treats graph generation as a sequential process. It generates nodes one at a time and for each new node, it decides its connections to previously generated nodes. The model consists of two main components: node-level RNN and edge-level RNN.
Architecture
Graph with node ordering can be uniquely mapped into a sequence of node and edge additions . The sequence has two levels: node-level and edge-level. Node-level sequence adds nodes one at the time, and each node-level step is an edge-level sequence. Edge-level sequence adds edges between existing nodes. where is the entire graph sequence, and is -th node-level step or edge-level sequence. where is -th edge-level step in -th node-level step.
The sequence is created from the Adjacency Matrix of a Graph.
Node-Level RNN
At each step, node-level RNN outputs a hidden state that summarizes the graph generated so far. The hidden state is used to initialize the edge-level RNN.
Edge-Level RNN
Edge-level RNN sequentially predict if the new node will connect to each of the previously generated nodes.
Link to original
More Expressive GNNs
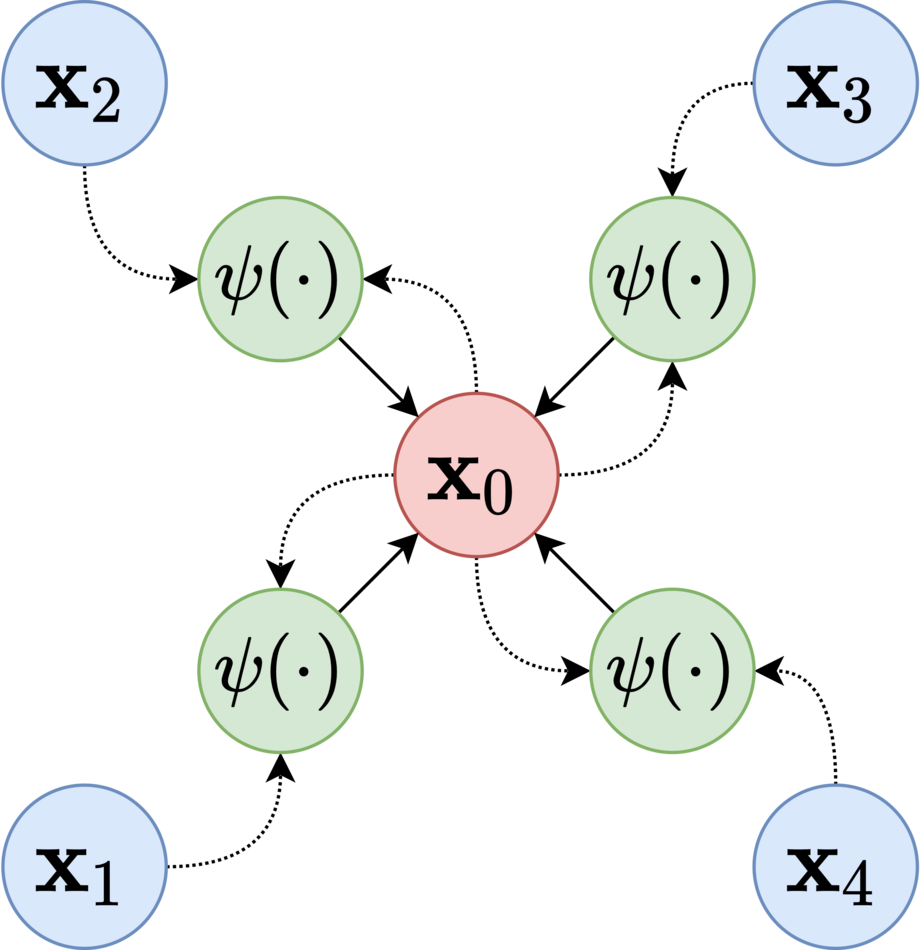
Position-Aware GNN
Definition
Position-aware GNN (P-GNN) is designed to compute position-aware Node Embedding.
Architecture
Effective Node Embedding should be able to learn to distinguish nodes and . However, standard GNN is not able to classify nodes and into different classes based on the network structure alone because the two nodes are symmetric/isomorphic in the graph, and their GNN rooted subtrees used for message aggregation are the same.
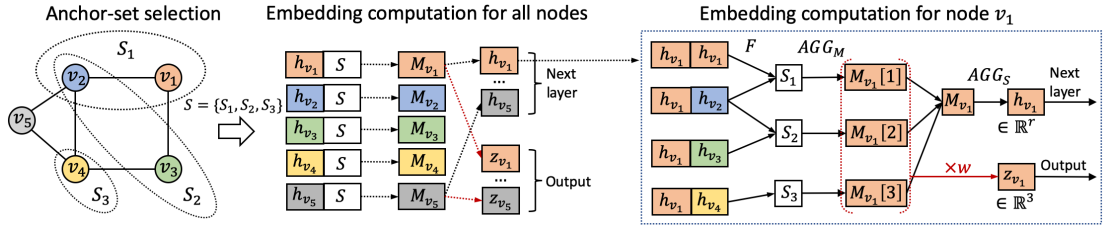
Anchor-Set
By using the distance of a given target node to each anchor-set as an augmented node feature, P-GNN can deal with the position-aware tasks. Where anchor-sets are randomly constructed.
P-GNN first samples sets of anchor nodes, computes the distance of a given target node to each anchor-set, and then learns a permutation-invariant aggregation over the anchor-sets. The model can capture positions/locations of nodes with respect to the anchor nodes.
Link to original
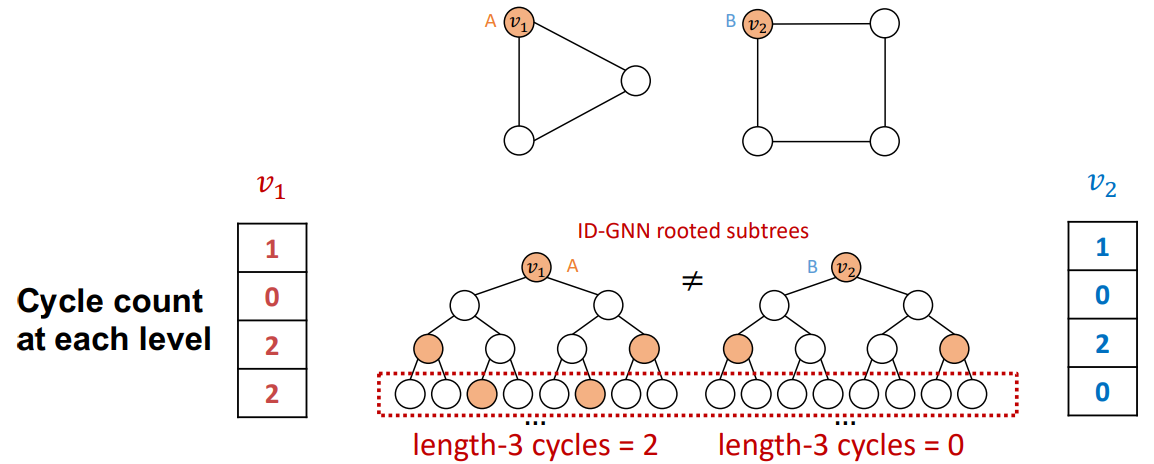
Identity-Aware GNN
Definition
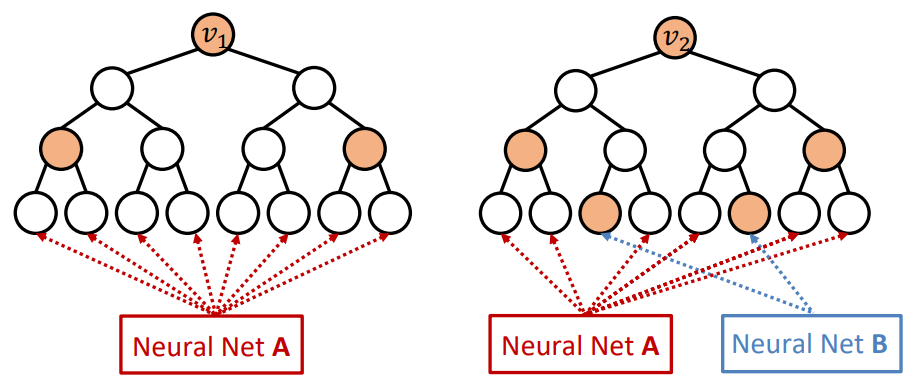
Identity-aware GNN (ID-GNN) is designed to compute structure-aware Node Embedding.
Architecture
Across all example tasks, traditional GNN will always assign the same embedding to both nodes, edges and graphs, because for all tasks the computational graphs are identical. In contrast, the colored computational graphs provided by ID-GNN allow for clear differentiation between the nodes of label A and label B, as the colored computational graphs are no longer identical across the tasks.
Heterogeneous Message Passing
ID-GNN implies inductive-node coloring by applying different message/aggregation to nodes with different colorings.
To embed a node , extract -centered local network (ego network) and assign a unique coloring to the central node of the network. The message passing and aggregation differ by the node’s coloring.
ID-GNN-Fast
The ID-GNN model can be simplified by just adding the cycle count at each level as an augmented node feature without using the two different networks.
Link to original
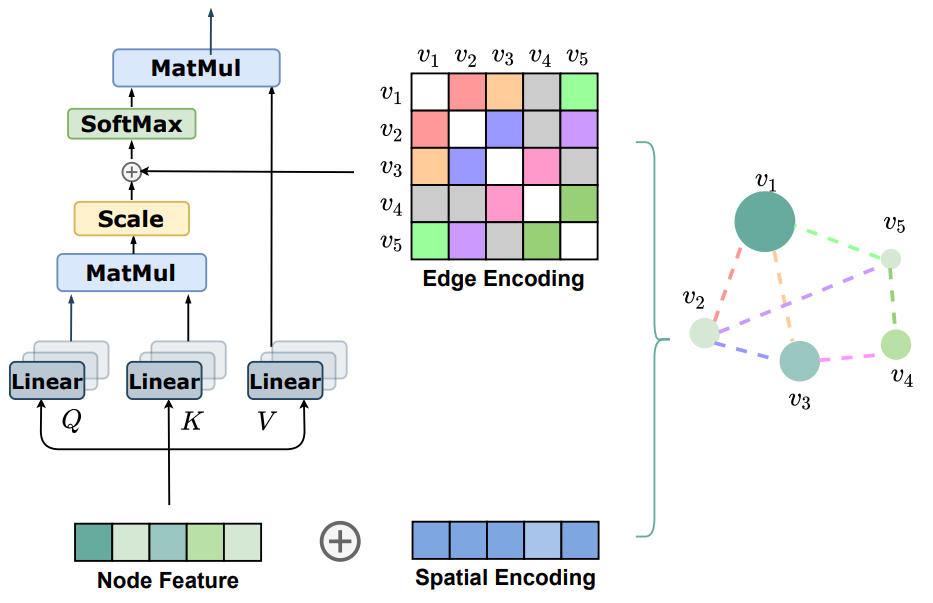
Graph Transformers
Transformer-Based GNN
Definition
Transformer-based GNN address some of the limitations of traditional GNNs (such as cycle counting) while leveraging the Transformer architecture for processing Graph-structured data.
Architecture
Node Embedding
Each node is embedded into a vector using their features.
Positional Encoding
Unlike in standard Transformer where positional encodings represent sequential order, in graph scenarios, these encodings are used to capture the structural information of the graph. It is constructed from the Adjacency Matrix of the graph.
There exists various approaches constructing positional encoding vectors.
Relative Distance
This method directly using the idea of Position-Aware GNN. The distance of a given target node to each anchor-set as a positional encoding.
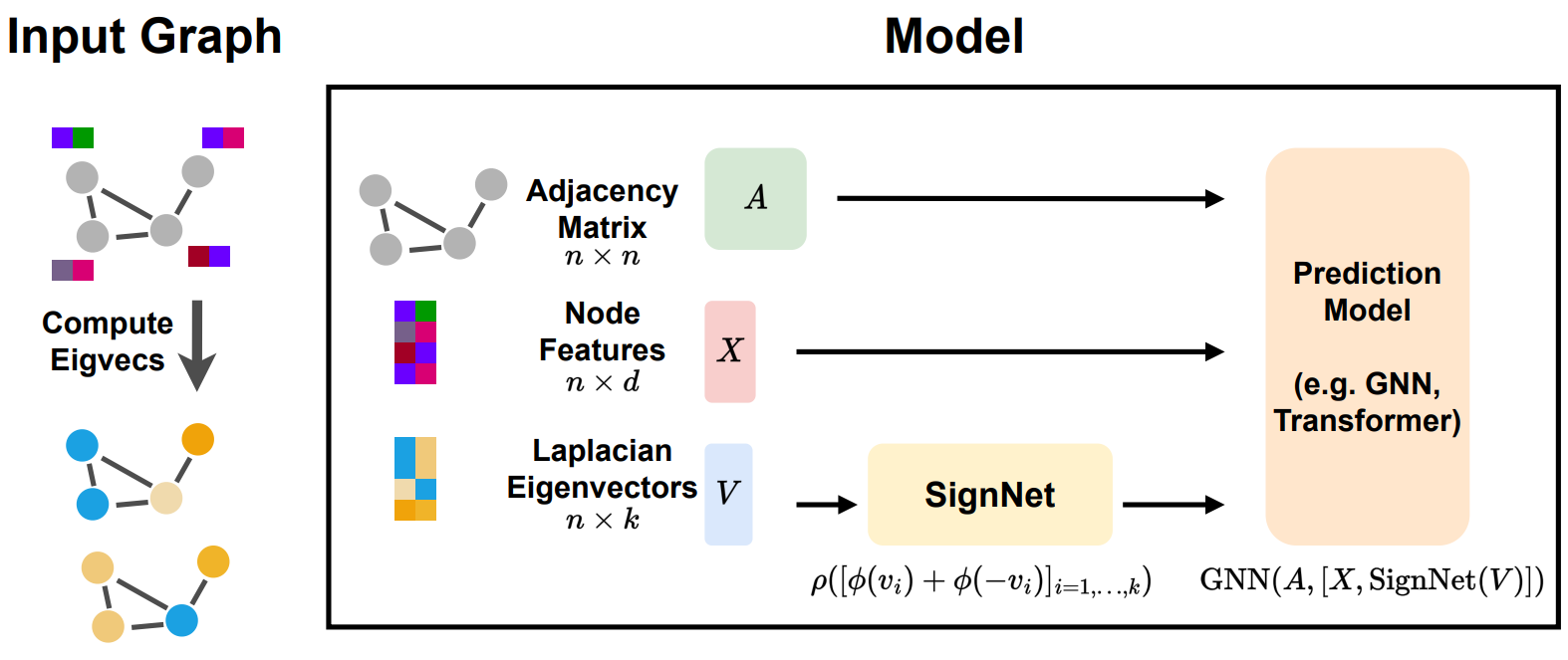
Laplacian Eigenvectors
Calculate the eigenvector matrix of the Laplacian Matrix, and use each row of the eigenvector matrix as a positional encoding of each node.
The signs of the eigenvectors may change model’s prediction. SigNet uses a neural network to get sign-invariant positional encoding to prevent this problem. where is the -th Eigenvector of the Laplacian Matrix, and and are neural network (MLP, GNN, etc.)
Self Attention
The edge features are used for adjusting the attention weights of the nodes.
If there is edge between nodes and with features , it is linearly transformed If there is no edge, find the shortest edge path between and , and define Then, it is added to the corresponding attention weight. where are learnable parameters.
Link to original
GNNs for Large Graphs
Neighbor Sampling
Definition
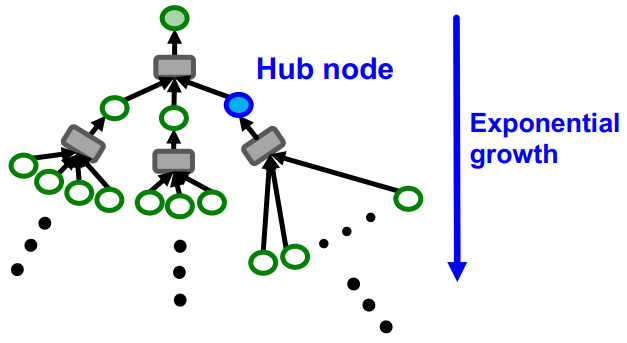
Neighbor sampling is used to efficiently compute node embeddings in large graphs where considering the entire neighborhood of a node becomes computationally expensive. Instead of using the entire -hop neighborhood of a node to compute its embedding, we sample a subset of neighbors at each hop. It is used for GraphSAGE model.
Constructing the exact computational graph for each node is computationally heavy. The computation graph becomes exponentially large with respect to the layer size , and explodes when it hits a hub (high-degree) node. For this reason, the neighbor sampling randomly samples at most neighbors at each hop.
Algorithm
- Randomly sample root nodes, where to the total number of nodes in the graph.
- For each sampled root node , construct the computational graph by randomly sampling at most neighbors at each hop.
- Update the embeddings of the root nodes.
Limitations
Link to original
- Smaller leads to more efficient neighborhood aggregation, but results are less stable due to the large variance in neighbor aggregation.
- Even with neighbor sampling, the size of the computational graph is still exponential with respect to number of GNN layers
- Random sampling is fast, but it may sample unimportant nodes.
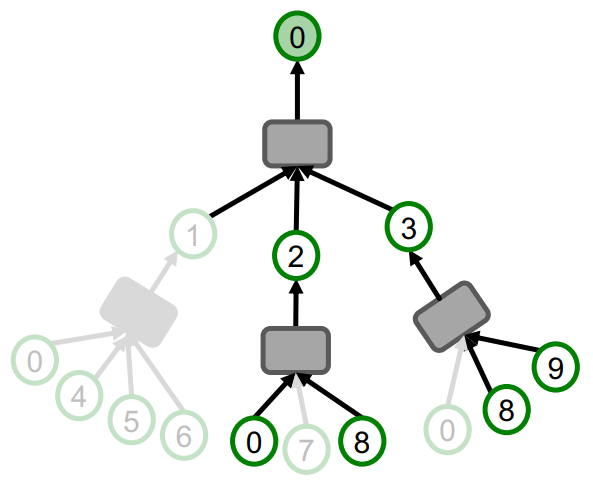
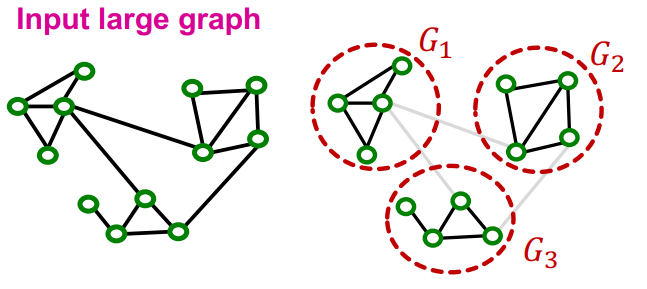
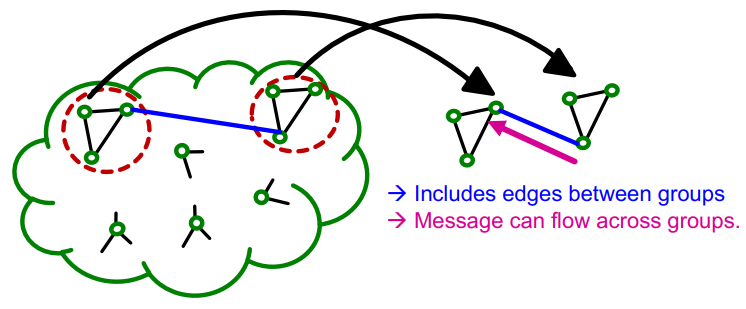
Cluster-GCN
Definition
Cluster-GCN is an algorithm for training GCN on large-scale graphs. The main idea of Cluster-GCN is to address the computational redundancy occurring in Neighbor Sampling, when nodes in a mini-batch share many neighbors.
Instead of sampling individual nodes or neighbors, Cluster-GCN samples small subgraphs from the large graph. It then performs efficient layer-wise node embedding updates over these subgraphs, where the subgraphs should retain edge connectivity structure of the original graph as much as possible.
If only single group is used per minibatch, the induced subgraph removes between-group links and the graph community detection algorithm puts similar nodes together in the same group. So the sampled nodes are not diverse enough to represent the entire graph structure. For this reason, the cluster-GCN aggregate multiple node groups per mini-batch.
Algorithm
Link to original
- The given graph is partitioned into groups of nodes (subgraphs) using community detection algorithms such as Louvain or METIS algorithm.
- For each mini-batch, randomly sample multiple node groups and construct node-induced subgraph of the aggregated node group.
- Update the embeddings of the nodes of the subgraph.
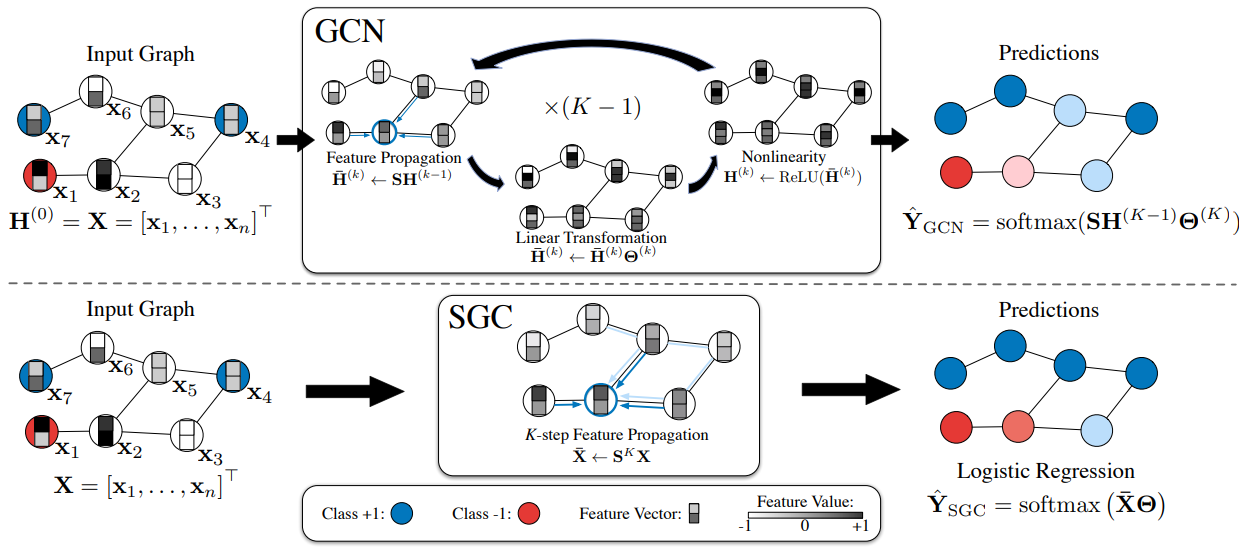
Simple Graph Convolution
Definition
Simple graph convolution (SGC) is a simplified variant of GCN. It simplifies GCN without losing much performance by removing the non-linear Activation Function. Due to the simplicity, the SGC can be more computationally efficient, especially for larger graphs. The simplification strategy is very similar to the one used by LightGCN for recommender systems.
The -hops simple graph convolution network for a classification task can be written in where:
- is the normalized adjacency matrix for the graph
- is the Adjacency Matrix of the graph.
- is the Degree Matrix of the matrix
- is the fixed input feature matrix of nodes.
- is the only weight matrix.
SGC doesn’t require building a computational graph or sample a subgraph, so can be applied for large graphs.
Link to original
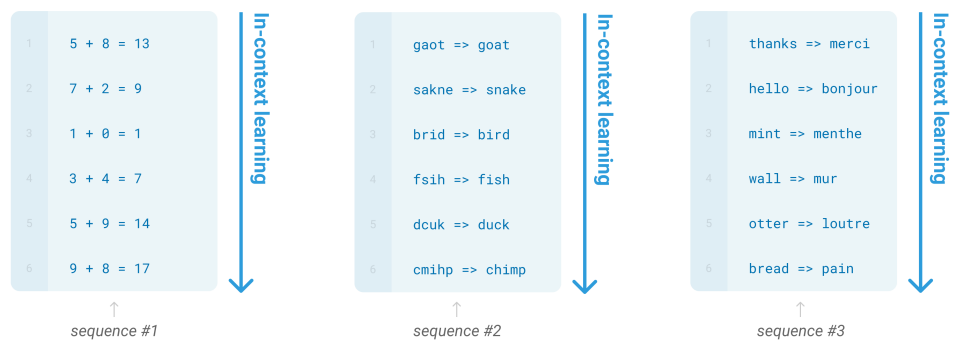
In-Context Learning Over Graphs
In-Context Learning
Definition
In-context learning (ICL) is a method utilizing pre-trained model to solve new tasks without fine-tuning by showing a few examples of the desire task to the model, allowing the model to infer the pattern and apply it to new instances. Unlike traditional machine learning, ICL doesn’t involve updating the model’s parameters. The model uses its existing knowledge to interpret and apply the new information.
Link to original
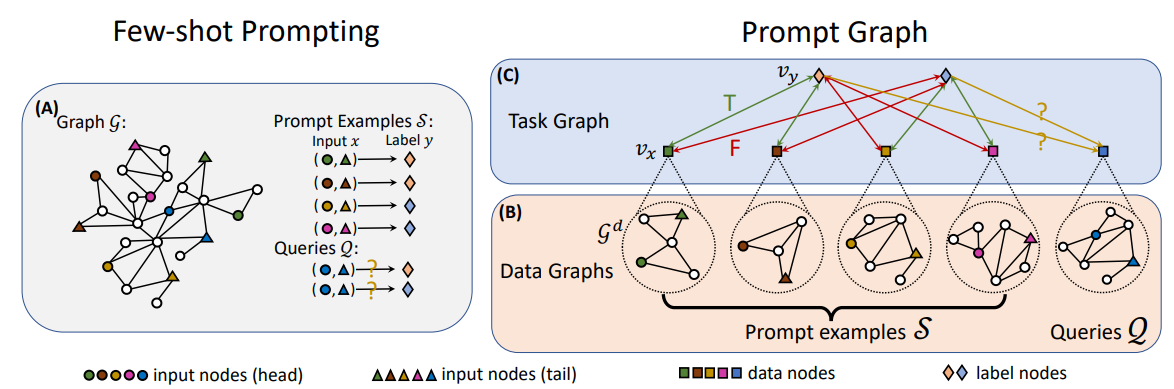
PRODIGY
Definition
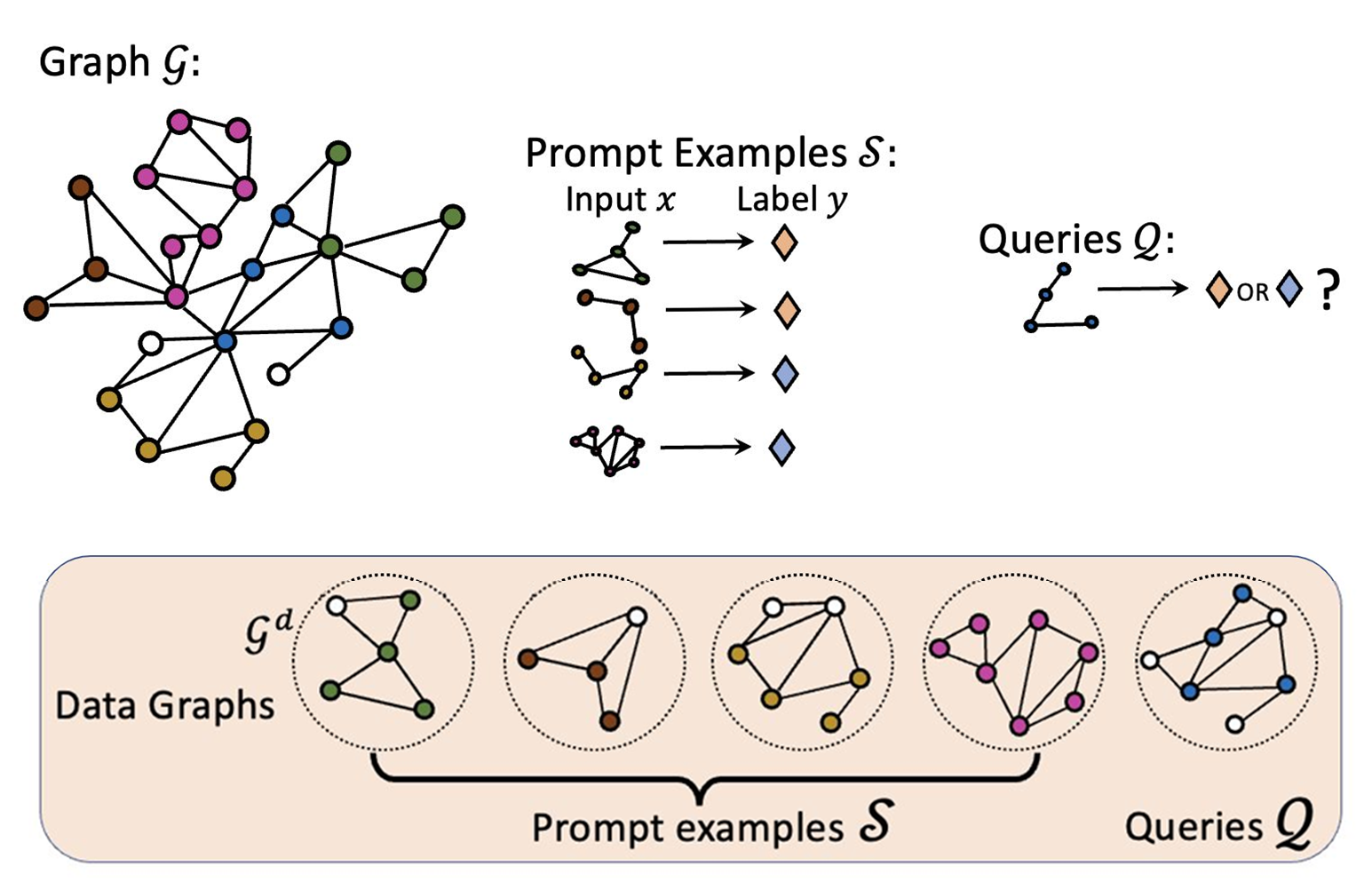
Pretraining Over Diverse In-Context Graph Systems (PRODIGY) model is a pretraining framework that enables In-Context Learning over graphs. The key idea of PRODIGY is to formulate in-context learning over graphs with a novel prompt graph representation. It is similar to the few-shot prompting widely used for LLM.
Architecture
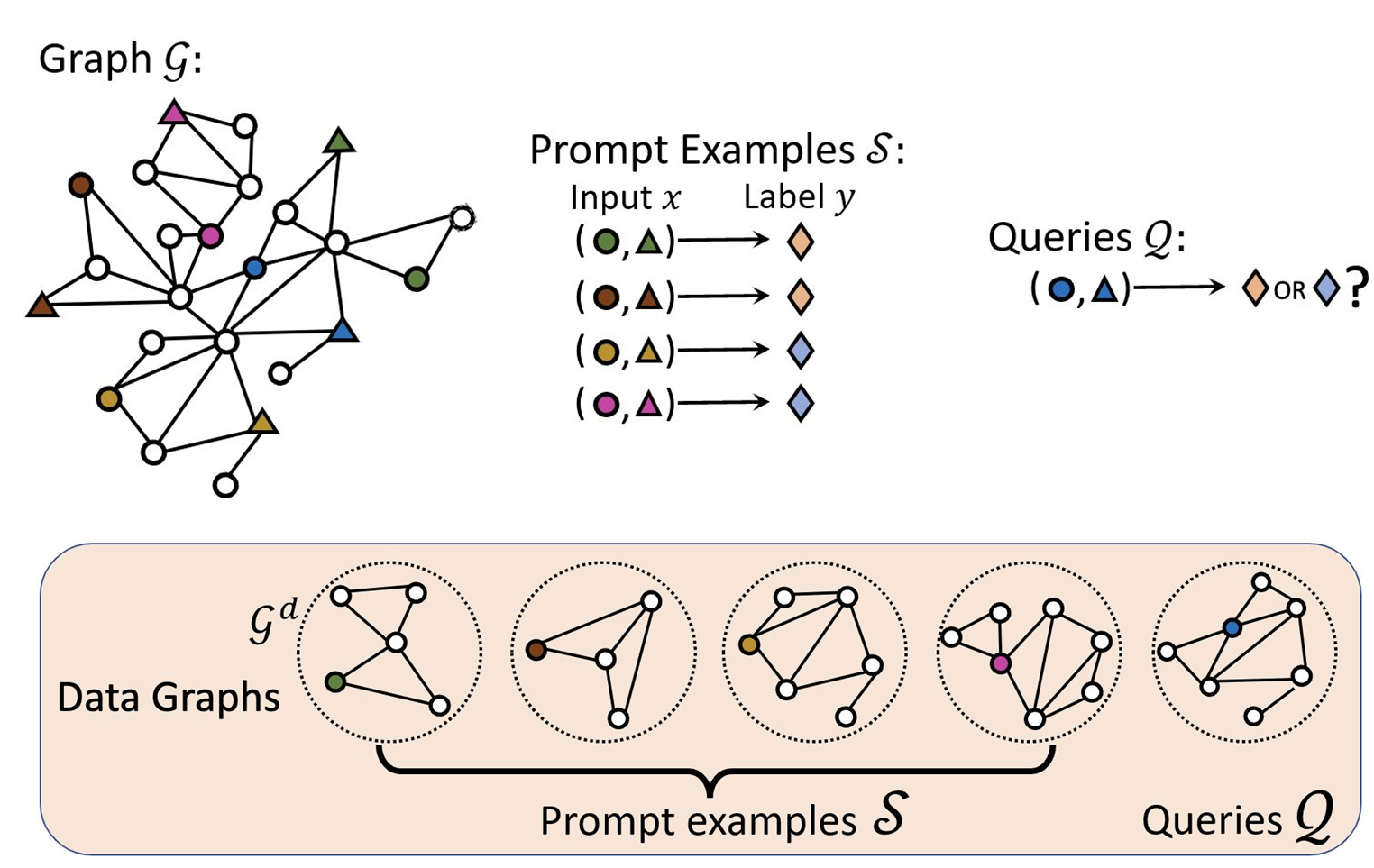
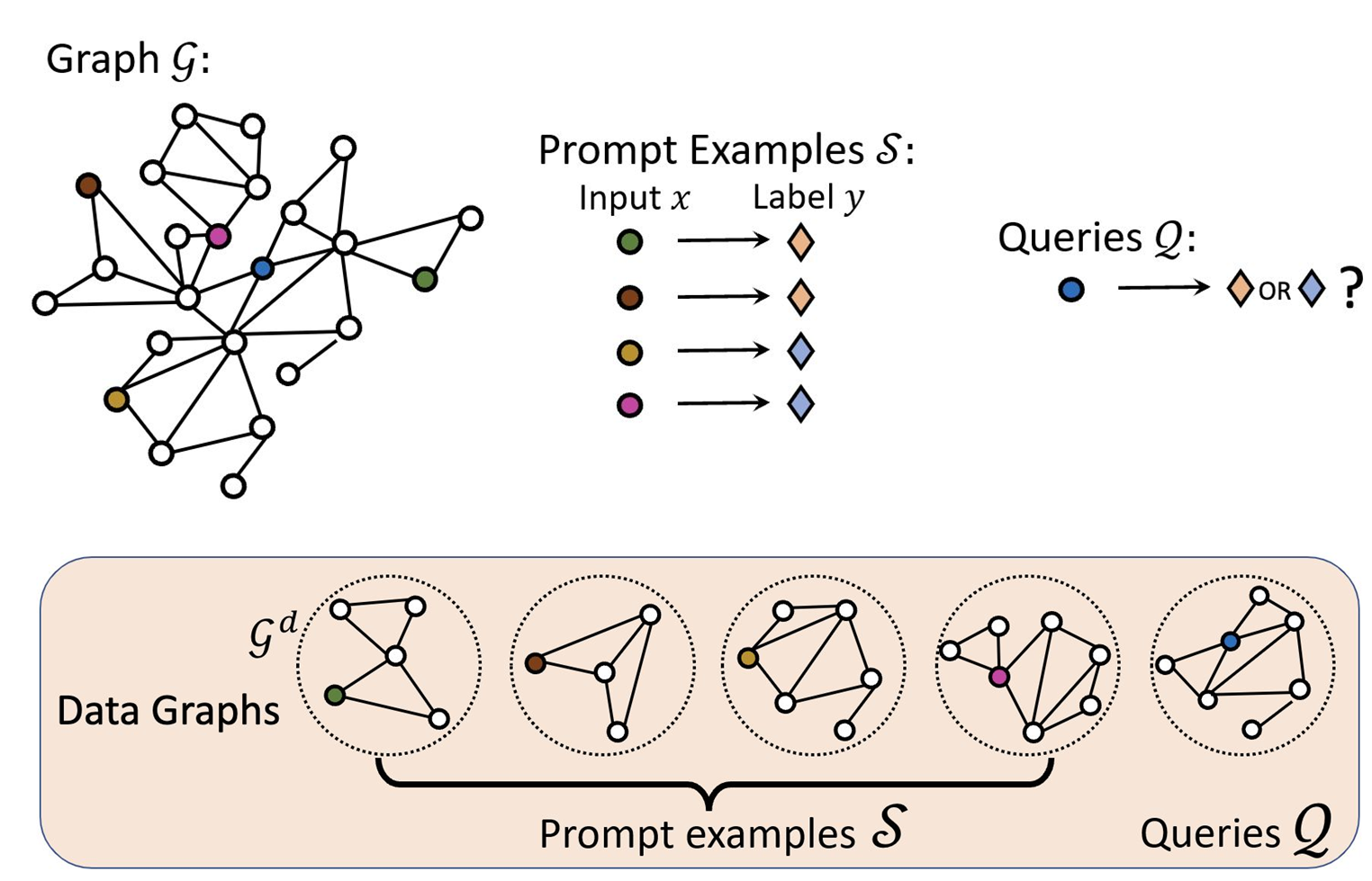
Prompt Graph
- Data graphs of the input nodes/edges/subgraphs are contextualized to by an embedding model such as GCN or GAT.
For node classification problem, the embedding of root node of the data graph is used as data node embedding. For link classification problem, the data node embedding is calculated using the edge’s nodes. where and are learnable parameters.
- Link Prediction
- Node Classification
- Graph Classification
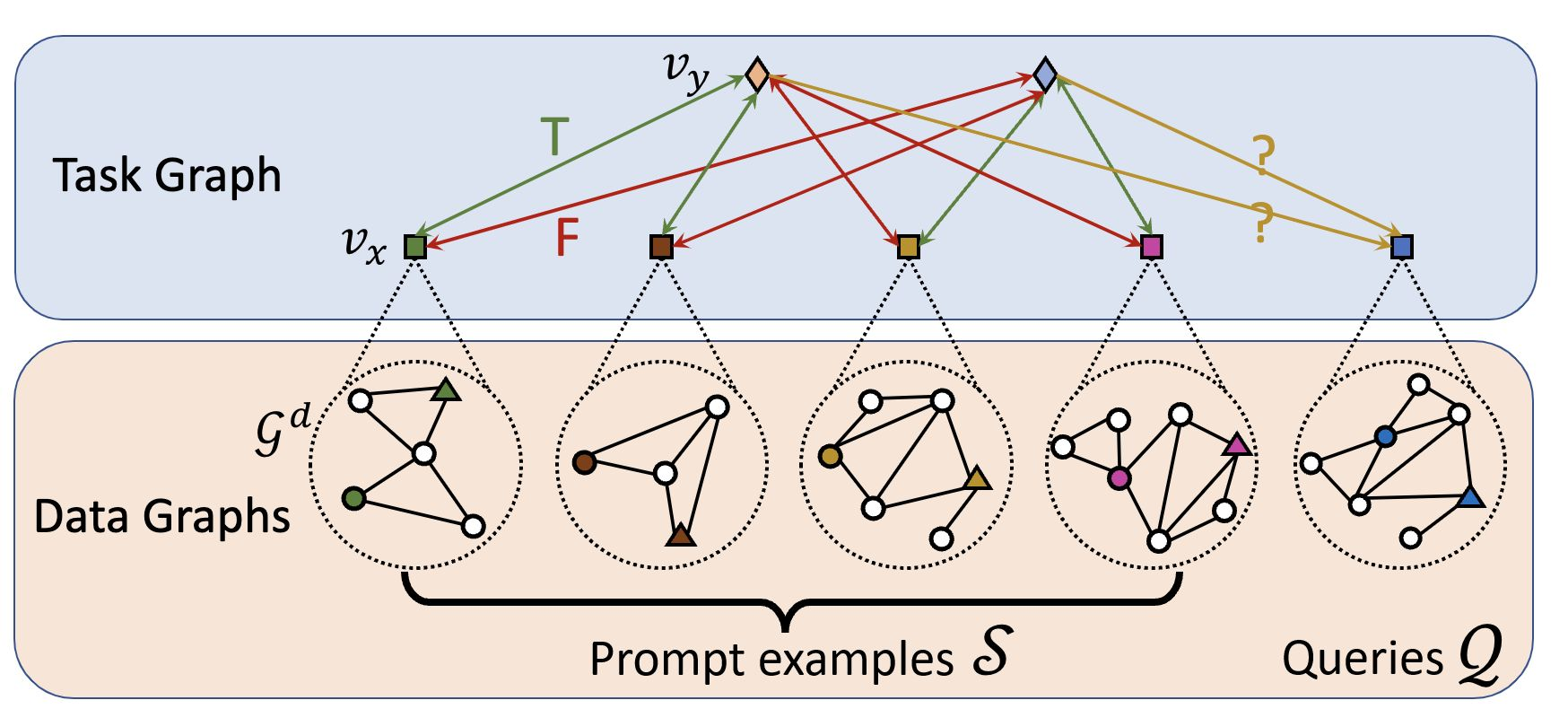
- Task graph is constructed using the contextualized data graphs and the labels. The edges between the data and label node groups are fully connected.
- The task graph is fed into the another GAT to obtain updated representation of data nodes and label nodes.
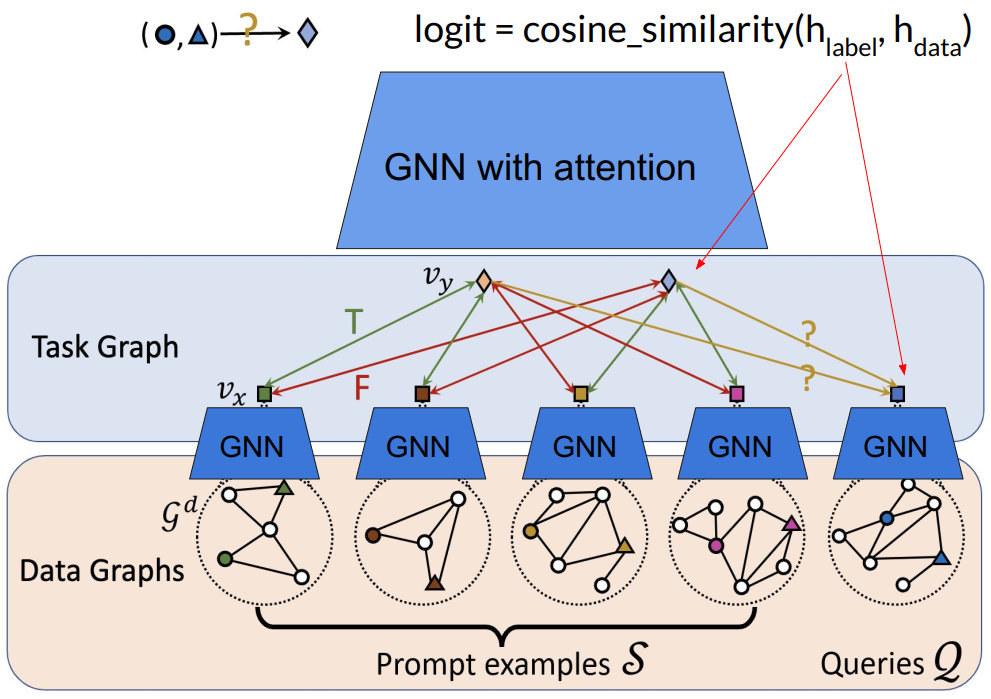
- The classification is performed by the cosine similarity of the embedded nodes of the target graph.
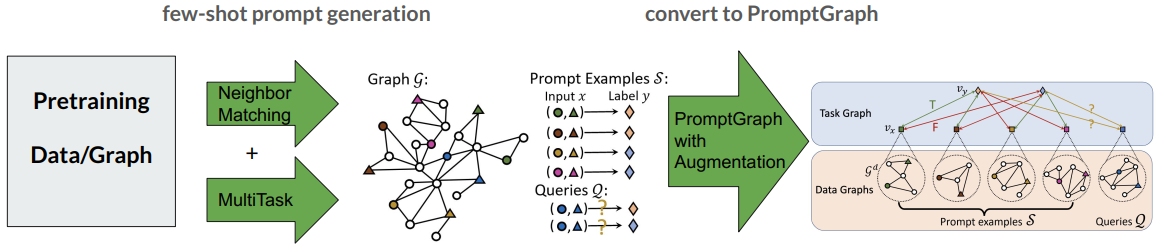
Pretraining
The PRODIGY model is trained by the neighbor matching task and the multi(edge/node/subgraph)task.
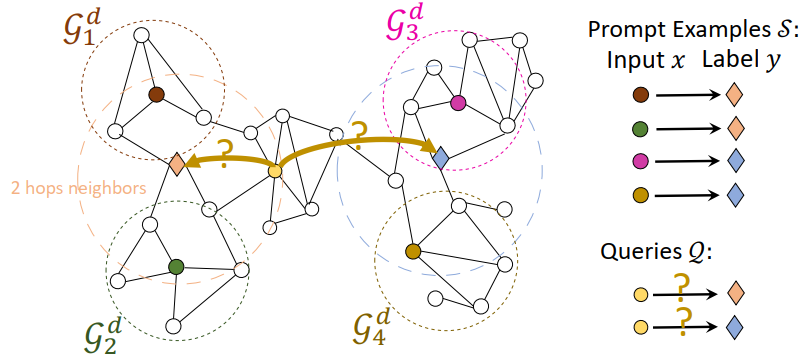
Neighbor Matching
We sample multiple subgraphs from the pretraining graph as the local neighborhoods, and we say a node belongs to a local neighborhood if it is in the sampled subgraph. The sampled subgraphs are used as the prompt/query data graphs
Multitask
In the pretraining stage, each data graph of prompts and queries is constructed by sampling -hop neighborhoods of the randomly sampled nodes.
Link to original