Definition
Neighbor sampling is used to efficiently compute node embeddings in large graphs where considering the entire neighborhood of a node becomes computationally expensive. Instead of using the entire -hop neighborhood of a node to compute its embedding, we sample a subset of neighbors at each hop. It is used for GraphSAGE model.
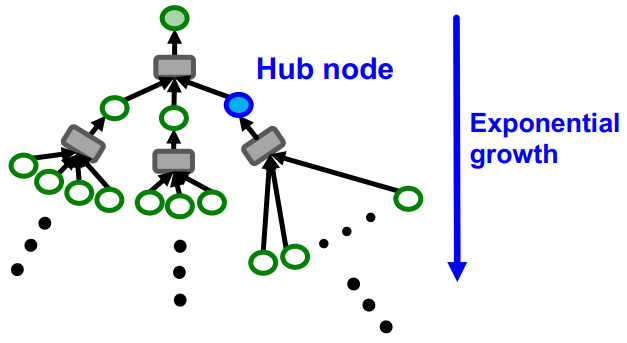
Constructing the exact computational graph for each node is computationally heavy. The computation graph becomes exponentially large with respect to the layer size , and explodes when it hits a hub (high-degree) node. For this reason, the neighbor sampling randomly samples at most neighbors at each hop.
Algorithm
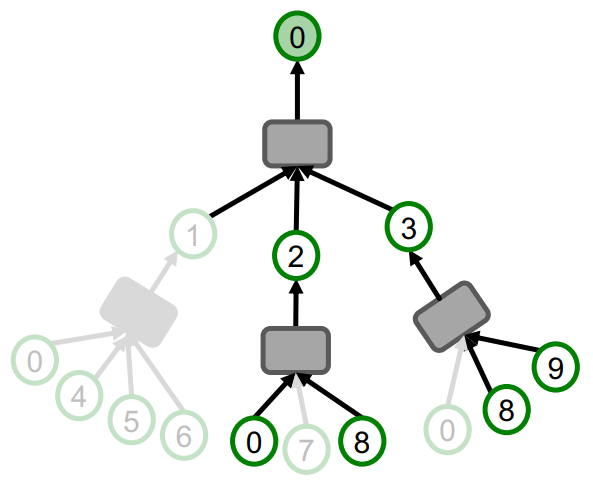
- Randomly sample root nodes, where to the total number of nodes in the graph.
- For each sampled root node , construct the computational graph by randomly sampling at most neighbors at each hop.
- Update the embeddings of the root nodes.
Limitations
- Smaller leads to more efficient neighborhood aggregation, but results are less stable due to the large variance in neighbor aggregation.
- Even with neighbor sampling, the size of the computational graph is still exponential with respect to number of GNN layers
- Random sampling is fast, but it may sample unimportant nodes.