Optimization
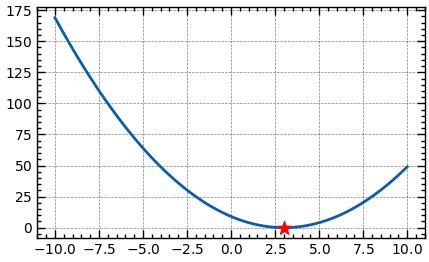
from scipy.optimize import minimize
f = lambda x: (x-3)**2
res = minimize(f, np.array([2]))
print(f'argmin: {res.x.item():.5f}, min: {res.fun:.5f}')Optimization with Constraints and Bounds
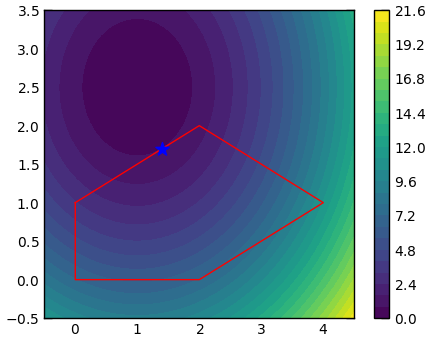
f = lambda x: (x[0] - 1) ** 2 + (x[1] - 2.5) ** 2
# inequality constraints should be a form of "eq >= 0".
constraints = [dict(type='ineq', fun=(lambda x: x[0] - 2 * x[1] + 2)),
dict(type='ineq', fun=(lambda x: -x[0] - 2 * x[1] + 6)),
dict(type='ineq', fun=(lambda x: -x[0] + 2 * x[1] + 2))]
bounds = [(0, None), (0, None)]
res = minimize(f, np.array([1, 2.5]), bounds=bounds, constraints=constraints)
print(f'argmin: {res.x}, min: {res.fun:.5f}')Interpolation
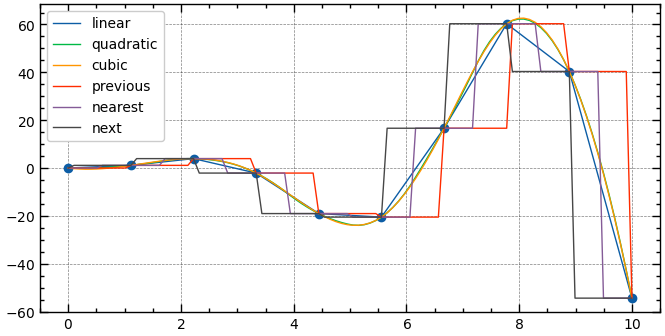
['linear', 'quadratic', 'cubic', 'previous', 'nearest', 'next']
from scipy.interpolate import interp1d
f = interp1d(x, y, kind='quadratic')
x_dense = np.linspace(0, 10, 100)
y_interp = f(x_dense) Curve Fitting
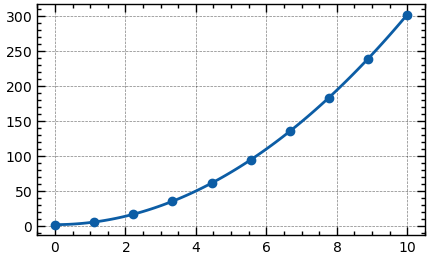
from scipy.optimize import curve_fit
x_data = np.linspace(0, 10, 10)
y_data = 3 * x_data ** 2 + 2
f = lambda x, a, b: a * x ** 2 + b
popt, _ = curve_fit(f, x_data, y_data, p0=[0])
x_dense = np.linspace(0, 10, 100)
y_dense = f(x_dense, *popt)Integration
from scipy.integrate import quad
integrand = lambda x: x ** 2 * np.sin(2 * x) * np.exp(-x)
integral, integral_error = quad(integrand, 0, 1) Multiple Integral
from scipy.integrate import dblquad
integrand = lambda y, x: np.sin(x + y ** 2)
lwr_y, upr_y = lambda x: -x, lambda x: x ** 2
integral, integral_error = dblquad(integrand,
0, 1,
lwr_y, upr_y)Differential Equation
ODE
from scipy.integrate import odeint
alpha, beta = 3, 5
dvdt = lambda v, t: alpha * v ** 2 - beta
v0 = 0
t_range = np.linspace(0, 1, 100)
sol = odeint(dvdt, v0, t_range)
v_range = sol.ravel()Coupled ODE
def dSdx(S, x):
y1, y2 = S
dy1dx = y1 + y2 ** 2 + 3 * x
dy2dx = 3 * y1 + y2 ** 3 - np.cos(x)
return dy1dx, dy2dx
y1_0, y2_0 = 0, 0
S_0 = (y1_0, y2_0)
x_range = np.linspace(0, 1, 100)
sol = odeint(dSdx, S_0, x_range)
y1, y2 = sol.T Second Order ODE
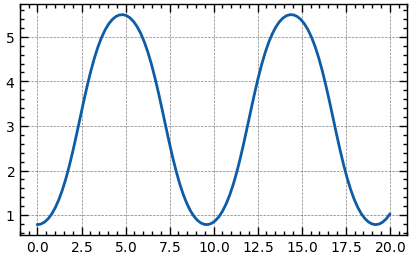
Transform the higher order ODE to the coupled first order ODEs ()
def dSdx(S, t):
omega, theta = S
d_omega = np.sin(theta)
d_theta = omega
return d_omega, d_theta
omega0 = 0
theta0 = np.pi / 4
S0 = (omega0, theta0)
t_range = np.linspace(0, 20, 100)
sol = odeint(dSdx, S0, t_range)
omega, theta = sol.TFourier Transform
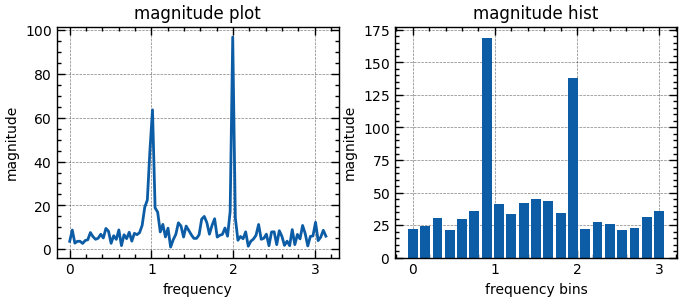
from scipy.fft import fft, fftfreq
t = np.linspace(0., 10 * np.pi, 200)
x = np.sin(2 * np.pi * t) + np.sin(4 * np.pi * t) + 0.5*np.random.randn(len(t))
N = len(x)
y = fft(x)[:N // 2]
f = fftfreq(N, t[1] - t[0])[:N // 2]
_, axs = plt.subplots(1, 2, figsize=(8, 3))
axs[0].plot(f, np.abs(y))
axs[0].set(title='magnitude plot', xlabel='frequency', ylabel='magnitude')
bin_size = 0.15
bins = np.arange(0, f.max() + bin_size, bin_size)
axs[1].hist(f, bins=bins, weights=np.abs(y), align='left', rwidth=0.8)
axs[1].set(title='magnitude hist', xlabel='frequency bins', ylabel='magnitude')
plt.show()Fourier Transform with Zero-Padding
By using zero-padding, we can get smoother spectral plots
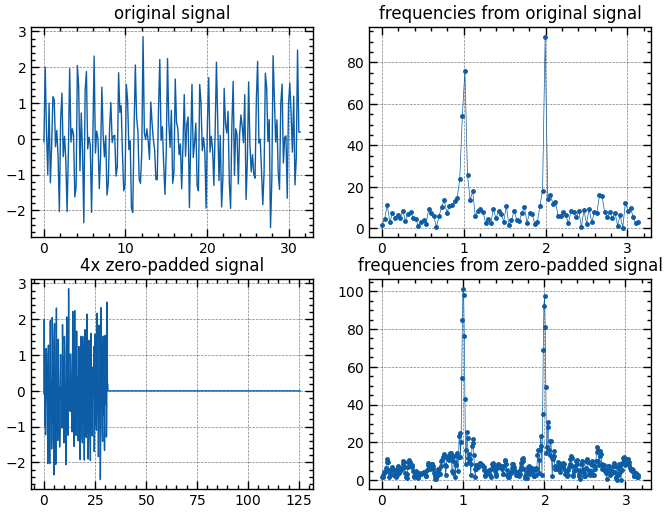
from scipy.fft import fft, fftfreq
_, axs = plt.subplots(2, 2, figsize=(8, 6))
t = np.linspace(0., 10 * np.pi, 200)
x = np.sin(2 * np.pi * t) + np.sin(4 * np.pi * t) + 0.5*np.random.randn(len(t))
N = len(x)
y = fft(x)[:N // 2]
f = fftfreq(N, t[1] - t[0])[:N // 2]
axs[0][0].plot(t, x, lw=1)
axs[0][0].set(title='original signal')
axs[0][1].plot(f, np.abs(y),'.-', markersize=5, lw=0.5)
axs[0][1].set(title='frequencies from original signal')
pad_ratio = 4
N *= pad_ratio
y = fft(x, n=N)[:N // 2]
f = fftfreq(N, t[1] - t[0])[:N // 2]
t = np.linspace(0., t.max()*pad_ratio, N)
x = np.concat([x, np.zeros(N-len(x))])
axs[1][0].plot(t, x, lw=1)
axs[1][0].set(title='4x zero-padded signal')
axs[1][1].plot(f, np.abs(y),'.-', markersize=5, lw=0.5)
axs[1][1].set(title='frequencies from zero-padded signal')
plt.show()Statistics
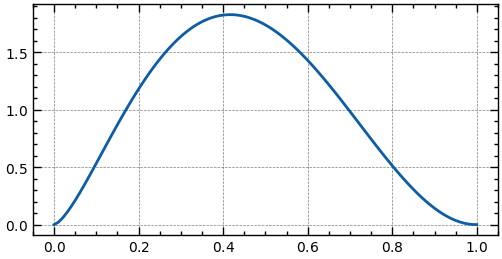
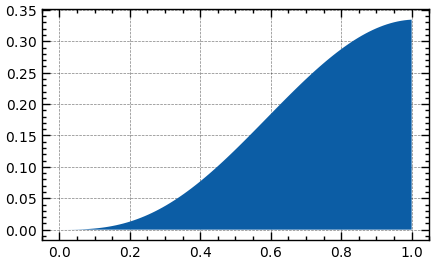
from scipy.stats import beta
a, b = 2.5, 3.1
mean, var, skew, kurt = beta.stats(a, b, moments='mvsk') # Get the moments of the distribution
print(f'{mean=:.3f}, {var=:.3f}, {skew=:.3f}, {kurt=:.3f}')
_, ax = plt.subplots(figsize=(6, 3))
x_range = np.linspace(0, 1, 100)
ax.plot(x_range, beta.pdf(x_range, a, b)) # Plot the PDF
plt.show()Custom Distribution
import scipy.stats as st
# Define a custom distribution
class mr_p_solver_dist(st.rv_continuous):
def _pdf(self,x, a1, a2, b1, b2):
return 1/(2*(a1*b1+a2*b2))*(b1*np.exp(-np.sqrt(x/a1)) + b2*np.exp(-np.sqrt(x/a2)))
my_rv = mr_p_solver_dist(a=0, b=np.inf)
# Random Sampling
print(my_rv.rvs(a1, a2, b1, b2, size=10))
_, ax = plt.subplots(figsize=(5, 3))
a1, a2, b1, b2 = 2, 3, 1, 2
x = np.linspace(my_rv.ppf(0.01, a1, a2, b1, b2), my_rv.ppf(0.99, a1, a2, b1, b2), 100)
y = my_rv.pdf(x, a1, a2, b1, b2)
ax.plot(x, y)
ax.set(title='Custom PDF', yscale='log')
plt.show()